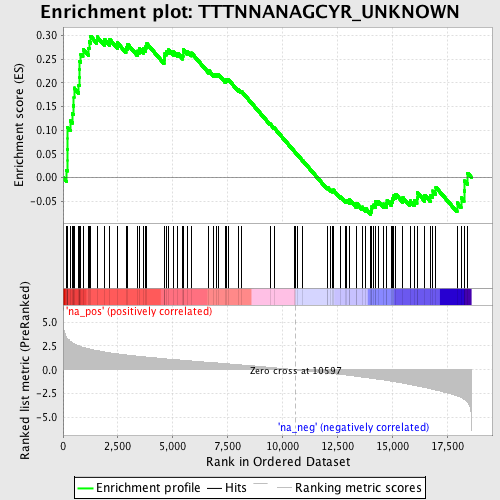
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTTNNANAGCYR_UNKNOWN |
| Enrichment Score (ES) | 0.2988566 |
| Normalized Enrichment Score (NES) | 1.3786774 |
| Nominal p-value | 0.03168317 |
| FDR q-value | 0.91768277 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HIST1H2AB | 148 | 3.391 | 0.0160 | Yes | ||
| 2 | HIST1H1A | 188 | 3.292 | 0.0372 | Yes | ||
| 3 | HIST1H3C | 190 | 3.285 | 0.0603 | Yes | ||
| 4 | HIST1H3D | 200 | 3.261 | 0.0829 | Yes | ||
| 5 | HIST2H2BE | 202 | 3.254 | 0.1059 | Yes | ||
| 6 | HIST1H3B | 329 | 2.978 | 0.1201 | Yes | ||
| 7 | HIST1H3H | 423 | 2.820 | 0.1351 | Yes | ||
| 8 | HIST1H2BH | 465 | 2.770 | 0.1524 | Yes | ||
| 9 | HIST1H2BL | 494 | 2.737 | 0.1703 | Yes | ||
| 10 | HIST1H1B | 499 | 2.734 | 0.1894 | Yes | ||
| 11 | HIST1H2BF | 714 | 2.498 | 0.1955 | Yes | ||
| 12 | HIST1H2BN | 745 | 2.472 | 0.2114 | Yes | ||
| 13 | HIST1H2BJ | 752 | 2.467 | 0.2285 | Yes | ||
| 14 | DPT | 759 | 2.464 | 0.2456 | Yes | ||
| 15 | HIST1H2BB | 808 | 2.429 | 0.2602 | Yes | ||
| 16 | FBXO21 | 907 | 2.345 | 0.2715 | Yes | ||
| 17 | HIST1H2BK | 1154 | 2.189 | 0.2736 | Yes | ||
| 18 | HIST1H2BG | 1190 | 2.169 | 0.2871 | Yes | ||
| 19 | HIST1H4A | 1253 | 2.136 | 0.2989 | Yes | ||
| 20 | CLDN10 | 1544 | 2.005 | 0.2974 | No | ||
| 21 | SLC35A2 | 1892 | 1.864 | 0.2918 | No | ||
| 22 | HIST1H2AE | 2129 | 1.775 | 0.2916 | No | ||
| 23 | HIST1H2BA | 2474 | 1.667 | 0.2848 | No | ||
| 24 | RGN | 2868 | 1.556 | 0.2746 | No | ||
| 25 | INPPL1 | 2937 | 1.538 | 0.2818 | No | ||
| 26 | HIST1H1E | 3390 | 1.421 | 0.2675 | No | ||
| 27 | HIST1H3F | 3475 | 1.403 | 0.2729 | No | ||
| 28 | BLNK | 3662 | 1.362 | 0.2724 | No | ||
| 29 | LHX2 | 3769 | 1.335 | 0.2762 | No | ||
| 30 | ETS1 | 3817 | 1.325 | 0.2830 | No | ||
| 31 | SMO | 4602 | 1.155 | 0.2488 | No | ||
| 32 | OLR1 | 4613 | 1.153 | 0.2564 | No | ||
| 33 | HIST1H2BC | 4639 | 1.148 | 0.2632 | No | ||
| 34 | TBXAS1 | 4720 | 1.132 | 0.2669 | No | ||
| 35 | FGD4 | 4810 | 1.114 | 0.2700 | No | ||
| 36 | MTSS1 | 5028 | 1.076 | 0.2658 | No | ||
| 37 | HIST1H1T | 5230 | 1.036 | 0.2623 | No | ||
| 38 | BMPR2 | 5454 | 0.992 | 0.2573 | No | ||
| 39 | ONECUT2 | 5468 | 0.988 | 0.2636 | No | ||
| 40 | MYH2 | 5472 | 0.988 | 0.2704 | No | ||
| 41 | MEIS1 | 5679 | 0.952 | 0.2660 | No | ||
| 42 | CDH16 | 5850 | 0.920 | 0.2633 | No | ||
| 43 | HIST1H2AI | 6643 | 0.775 | 0.2260 | No | ||
| 44 | TCERG1L | 6875 | 0.731 | 0.2187 | No | ||
| 45 | DNAJB8 | 6980 | 0.718 | 0.2182 | No | ||
| 46 | GPM6B | 7073 | 0.702 | 0.2182 | No | ||
| 47 | KCNK12 | 7404 | 0.645 | 0.2049 | No | ||
| 48 | SERPINA7 | 7435 | 0.639 | 0.2078 | No | ||
| 49 | CNN1 | 7538 | 0.619 | 0.2067 | No | ||
| 50 | HIST1H2AC | 7994 | 0.532 | 0.1859 | No | ||
| 51 | NAP1L5 | 8117 | 0.508 | 0.1829 | No | ||
| 52 | NDNL2 | 9434 | 0.246 | 0.1136 | No | ||
| 53 | KCNIP4 | 9616 | 0.207 | 0.1053 | No | ||
| 54 | HIST1H2AH | 10527 | 0.016 | 0.0562 | No | ||
| 55 | HIST3H2BB | 10574 | 0.003 | 0.0538 | No | ||
| 56 | SPRED1 | 10692 | -0.025 | 0.0476 | No | ||
| 57 | HRASLS | 10931 | -0.079 | 0.0353 | No | ||
| 58 | HIST2H2AC | 12033 | -0.332 | -0.0218 | No | ||
| 59 | LIN28 | 12044 | -0.335 | -0.0200 | No | ||
| 60 | HIST1H1C | 12205 | -0.371 | -0.0260 | No | ||
| 61 | DHRS3 | 12292 | -0.394 | -0.0278 | No | ||
| 62 | NRAP | 12308 | -0.397 | -0.0258 | No | ||
| 63 | FOXP1 | 12643 | -0.483 | -0.0404 | No | ||
| 64 | CLDN2 | 12885 | -0.541 | -0.0496 | No | ||
| 65 | E2F3 | 12935 | -0.554 | -0.0484 | No | ||
| 66 | PPP3CA | 13049 | -0.589 | -0.0503 | No | ||
| 67 | HIST1H2AA | 13056 | -0.592 | -0.0464 | No | ||
| 68 | CENTD1 | 13377 | -0.685 | -0.0589 | No | ||
| 69 | SLC12A2 | 13380 | -0.685 | -0.0541 | No | ||
| 70 | HOXB6 | 13625 | -0.760 | -0.0619 | No | ||
| 71 | STARD13 | 13801 | -0.811 | -0.0656 | No | ||
| 72 | OTP | 14027 | -0.876 | -0.0716 | No | ||
| 73 | FGF17 | 14053 | -0.883 | -0.0667 | No | ||
| 74 | DSC1 | 14073 | -0.891 | -0.0614 | No | ||
| 75 | DGKG | 14124 | -0.910 | -0.0577 | No | ||
| 76 | AGTR2 | 14234 | -0.944 | -0.0569 | No | ||
| 77 | HIST1H2BE | 14239 | -0.945 | -0.0504 | No | ||
| 78 | SLC6A1 | 14357 | -0.983 | -0.0498 | No | ||
| 79 | PRKCQ | 14603 | -1.060 | -0.0555 | No | ||
| 80 | RANGAP1 | 14734 | -1.109 | -0.0547 | No | ||
| 81 | CAST | 14761 | -1.120 | -0.0482 | No | ||
| 82 | TCF4 | 14973 | -1.198 | -0.0511 | No | ||
| 83 | USP47 | 15015 | -1.213 | -0.0448 | No | ||
| 84 | AOC2 | 15061 | -1.230 | -0.0385 | No | ||
| 85 | HIST3H2A | 15160 | -1.272 | -0.0348 | No | ||
| 86 | HIST1H1D | 15486 | -1.401 | -0.0424 | No | ||
| 87 | SLC10A2 | 15810 | -1.536 | -0.0490 | No | ||
| 88 | SLC6A13 | 16004 | -1.629 | -0.0479 | No | ||
| 89 | MXI1 | 16130 | -1.684 | -0.0427 | No | ||
| 90 | CPNE1 | 16149 | -1.691 | -0.0318 | No | ||
| 91 | CYP51A1 | 16484 | -1.847 | -0.0367 | No | ||
| 92 | AMOT | 16756 | -1.994 | -0.0373 | No | ||
| 93 | NFATC1 | 16855 | -2.044 | -0.0281 | No | ||
| 94 | CHD4 | 16990 | -2.116 | -0.0204 | No | ||
| 95 | RNF44 | 17956 | -2.727 | -0.0532 | No | ||
| 96 | NDRG3 | 18139 | -2.908 | -0.0425 | No | ||
| 97 | GRK5 | 18273 | -3.087 | -0.0278 | No | ||
| 98 | ACN9 | 18295 | -3.125 | -0.0068 | No | ||
| 99 | HOXB4 | 18436 | -3.412 | 0.0097 | No |
