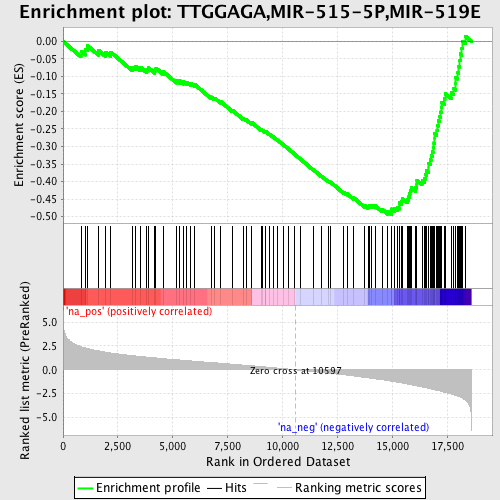
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | -0.49420226 |
| Normalized Enrichment Score (NES) | -2.3250597 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DGCR2 | 818 | 2.423 | -0.0274 | No | ||
| 2 | FGFR2 | 1028 | 2.265 | -0.0230 | No | ||
| 3 | CLSTN2 | 1102 | 2.222 | -0.0116 | No | ||
| 4 | EFNA3 | 1609 | 1.979 | -0.0252 | No | ||
| 5 | TGFBR2 | 1942 | 1.846 | -0.0304 | No | ||
| 6 | APLN | 2167 | 1.764 | -0.0303 | No | ||
| 7 | CSNK1G1 | 3152 | 1.482 | -0.0732 | No | ||
| 8 | ABCC3 | 3305 | 1.443 | -0.0714 | No | ||
| 9 | CLCF1 | 3521 | 1.393 | -0.0734 | No | ||
| 10 | ETS1 | 3817 | 1.325 | -0.0801 | No | ||
| 11 | GDI1 | 3898 | 1.305 | -0.0754 | No | ||
| 12 | FREQ | 4181 | 1.250 | -0.0820 | No | ||
| 13 | SOX30 | 4229 | 1.239 | -0.0760 | No | ||
| 14 | CPEB2 | 4567 | 1.163 | -0.0861 | No | ||
| 15 | ETV1 | 5172 | 1.046 | -0.1115 | No | ||
| 16 | MLLT6 | 5326 | 1.019 | -0.1127 | No | ||
| 17 | SNAP25 | 5493 | 0.983 | -0.1149 | No | ||
| 18 | ABCG4 | 5636 | 0.959 | -0.1159 | No | ||
| 19 | KCND2 | 5819 | 0.924 | -0.1193 | No | ||
| 20 | SLC4A4 | 5992 | 0.894 | -0.1225 | No | ||
| 21 | TNPO1 | 6758 | 0.754 | -0.1586 | No | ||
| 22 | MEF2D | 6907 | 0.727 | -0.1615 | No | ||
| 23 | FSTL1 | 7175 | 0.684 | -0.1712 | No | ||
| 24 | SYNGR1 | 7723 | 0.582 | -0.1967 | No | ||
| 25 | HOXA1 | 8231 | 0.487 | -0.2207 | No | ||
| 26 | OSBPL3 | 8344 | 0.466 | -0.2236 | No | ||
| 27 | NFIB | 8575 | 0.420 | -0.2331 | No | ||
| 28 | ACY1L2 | 8604 | 0.416 | -0.2317 | No | ||
| 29 | NFATC4 | 9023 | 0.339 | -0.2519 | No | ||
| 30 | NUP35 | 9086 | 0.325 | -0.2530 | No | ||
| 31 | SERTAD1 | 9221 | 0.294 | -0.2582 | No | ||
| 32 | DCX | 9246 | 0.290 | -0.2575 | No | ||
| 33 | ITSN1 | 9422 | 0.250 | -0.2653 | No | ||
| 34 | SGPL1 | 9574 | 0.217 | -0.2719 | No | ||
| 35 | ISL1 | 9792 | 0.168 | -0.2825 | No | ||
| 36 | TMEM47 | 10039 | 0.121 | -0.2949 | No | ||
| 37 | PDGFRA | 10254 | 0.075 | -0.3060 | No | ||
| 38 | SP4 | 10540 | 0.012 | -0.3213 | No | ||
| 39 | BTG2 | 10797 | -0.051 | -0.3348 | No | ||
| 40 | ANUBL1 | 11410 | -0.188 | -0.3665 | No | ||
| 41 | ETNK2 | 11786 | -0.272 | -0.3849 | No | ||
| 42 | OPHN1 | 12093 | -0.345 | -0.3990 | No | ||
| 43 | SCN8A | 12183 | -0.367 | -0.4013 | No | ||
| 44 | EMX2 | 12783 | -0.515 | -0.4301 | No | ||
| 45 | UBE2B | 12943 | -0.557 | -0.4348 | No | ||
| 46 | MTF2 | 13249 | -0.642 | -0.4468 | No | ||
| 47 | ZFP36L1 | 13742 | -0.797 | -0.4679 | No | ||
| 48 | BSN | 13899 | -0.834 | -0.4705 | No | ||
| 49 | KCTD7 | 13957 | -0.850 | -0.4677 | No | ||
| 50 | POM121 | 14062 | -0.886 | -0.4672 | No | ||
| 51 | SLC7A11 | 14220 | -0.939 | -0.4692 | No | ||
| 52 | PSMD11 | 14541 | -1.037 | -0.4793 | No | ||
| 53 | SOX12 | 14800 | -1.134 | -0.4854 | No | ||
| 54 | EPN2 | 14964 | -1.193 | -0.4859 | Yes | ||
| 55 | CAB39 | 14980 | -1.201 | -0.4784 | Yes | ||
| 56 | ARHGEF3 | 15107 | -1.252 | -0.4766 | Yes | ||
| 57 | CMTM6 | 15244 | -1.305 | -0.4749 | Yes | ||
| 58 | PLAGL2 | 15314 | -1.330 | -0.4694 | Yes | ||
| 59 | UBE2H | 15315 | -1.330 | -0.4602 | Yes | ||
| 60 | POU2F1 | 15430 | -1.374 | -0.4569 | Yes | ||
| 61 | YAF2 | 15453 | -1.388 | -0.4484 | Yes | ||
| 62 | IQGAP1 | 15693 | -1.489 | -0.4511 | Yes | ||
| 63 | SUMO2 | 15758 | -1.516 | -0.4440 | Yes | ||
| 64 | PUM1 | 15776 | -1.521 | -0.4344 | Yes | ||
| 65 | MOSPD2 | 15812 | -1.537 | -0.4257 | Yes | ||
| 66 | RPS6KA3 | 15856 | -1.562 | -0.4172 | Yes | ||
| 67 | ULK2 | 16069 | -1.657 | -0.4172 | Yes | ||
| 68 | ARRDC3 | 16120 | -1.680 | -0.4082 | Yes | ||
| 69 | DDX3X | 16124 | -1.681 | -0.3968 | Yes | ||
| 70 | RSBN1 | 16360 | -1.791 | -0.3971 | Yes | ||
| 71 | RDH11 | 16482 | -1.844 | -0.3908 | Yes | ||
| 72 | GABBR1 | 16526 | -1.869 | -0.3802 | Yes | ||
| 73 | YTHDF2 | 16562 | -1.887 | -0.3691 | Yes | ||
| 74 | ICK | 16656 | -1.935 | -0.3607 | Yes | ||
| 75 | HNRPUL1 | 16657 | -1.935 | -0.3473 | Yes | ||
| 76 | RAP1B | 16745 | -1.987 | -0.3382 | Yes | ||
| 77 | UHRF2 | 16772 | -2.003 | -0.3258 | Yes | ||
| 78 | NCOA1 | 16840 | -2.036 | -0.3153 | Yes | ||
| 79 | HIP2 | 16863 | -2.053 | -0.3023 | Yes | ||
| 80 | NOTCH2 | 16901 | -2.067 | -0.2900 | Yes | ||
| 81 | SLC9A2 | 16933 | -2.084 | -0.2772 | Yes | ||
| 82 | DDX3Y | 16936 | -2.085 | -0.2629 | Yes | ||
| 83 | UBAP2 | 17038 | -2.141 | -0.2536 | Yes | ||
| 84 | DONSON | 17071 | -2.156 | -0.2404 | Yes | ||
| 85 | MAPRE1 | 17095 | -2.168 | -0.2266 | Yes | ||
| 86 | ARID1A | 17156 | -2.208 | -0.2146 | Yes | ||
| 87 | RAP2C | 17200 | -2.232 | -0.2014 | Yes | ||
| 88 | ATRN | 17234 | -2.250 | -0.1877 | Yes | ||
| 89 | ATXN3 | 17252 | -2.261 | -0.1729 | Yes | ||
| 90 | CYR61 | 17381 | -2.338 | -0.1637 | Yes | ||
| 91 | TMEM2 | 17407 | -2.352 | -0.1487 | Yes | ||
| 92 | ZBTB1 | 17694 | -2.534 | -0.1467 | Yes | ||
| 93 | EPC2 | 17803 | -2.604 | -0.1345 | Yes | ||
| 94 | KPNA1 | 17888 | -2.676 | -0.1205 | Yes | ||
| 95 | MTHFD1L | 17895 | -2.683 | -0.1022 | Yes | ||
| 96 | RPP14 | 17991 | -2.757 | -0.0883 | Yes | ||
| 97 | MARCKS | 18036 | -2.799 | -0.0713 | Yes | ||
| 98 | FRAS1 | 18072 | -2.831 | -0.0536 | Yes | ||
| 99 | AP3S1 | 18099 | -2.860 | -0.0352 | Yes | ||
| 100 | RIOK3 | 18173 | -2.943 | -0.0188 | Yes | ||
| 101 | RBL2 | 18203 | -2.976 | 0.0002 | Yes | ||
| 102 | SFRS1 | 18333 | -3.183 | 0.0153 | Yes |
