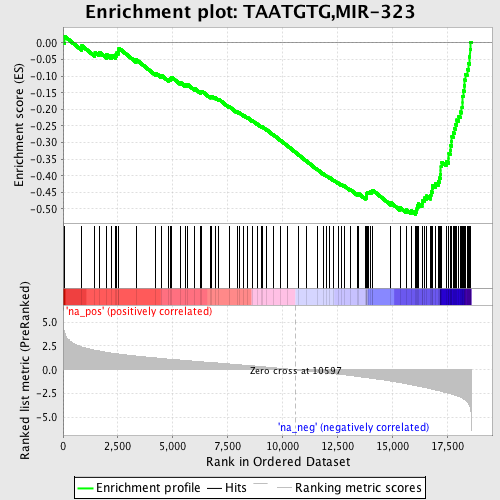
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | -0.51655895 |
| Normalized Enrichment Score (NES) | -2.4130526 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2B | 58 | 4.019 | 0.0204 | No | ||
| 2 | CREB1 | 854 | 2.389 | -0.0086 | No | ||
| 3 | FBN2 | 1452 | 2.042 | -0.0288 | No | ||
| 4 | SEMA6D | 1641 | 1.966 | -0.0275 | No | ||
| 5 | MCFD2 | 1985 | 1.829 | -0.0353 | No | ||
| 6 | CRK | 2219 | 1.743 | -0.0377 | No | ||
| 7 | PAK7 | 2378 | 1.693 | -0.0363 | No | ||
| 8 | EGR3 | 2449 | 1.673 | -0.0303 | No | ||
| 9 | PKP4 | 2513 | 1.659 | -0.0240 | No | ||
| 10 | SEPT3 | 2542 | 1.651 | -0.0158 | No | ||
| 11 | MAP3K3 | 3326 | 1.438 | -0.0497 | No | ||
| 12 | MYLIP | 4221 | 1.242 | -0.0907 | No | ||
| 13 | CDKN1B | 4465 | 1.185 | -0.0969 | No | ||
| 14 | FGD4 | 4810 | 1.114 | -0.1090 | No | ||
| 15 | HOXB5 | 4898 | 1.098 | -0.1072 | No | ||
| 16 | STAT5B | 4945 | 1.090 | -0.1033 | No | ||
| 17 | TMEM16D | 5340 | 1.015 | -0.1187 | No | ||
| 18 | CXCL12 | 5563 | 0.972 | -0.1250 | No | ||
| 19 | BET1 | 5671 | 0.953 | -0.1252 | No | ||
| 20 | SLC4A4 | 5992 | 0.894 | -0.1372 | No | ||
| 21 | GPM6A | 6271 | 0.842 | -0.1473 | No | ||
| 22 | DPF2 | 6327 | 0.833 | -0.1454 | No | ||
| 23 | EIF3S1 | 6709 | 0.762 | -0.1615 | No | ||
| 24 | TMOD1 | 6765 | 0.752 | -0.1601 | No | ||
| 25 | LPP | 6929 | 0.723 | -0.1647 | No | ||
| 26 | POU2F3 | 7100 | 0.699 | -0.1698 | No | ||
| 27 | NFKB1 | 7571 | 0.612 | -0.1916 | No | ||
| 28 | QSCN6L1 | 7926 | 0.543 | -0.2075 | No | ||
| 29 | TEAD1 | 8022 | 0.526 | -0.2096 | No | ||
| 30 | HOXA1 | 8231 | 0.487 | -0.2180 | No | ||
| 31 | FBN1 | 8415 | 0.453 | -0.2252 | No | ||
| 32 | PCDH9 | 8611 | 0.414 | -0.2333 | No | ||
| 33 | STK35 | 8850 | 0.373 | -0.2440 | No | ||
| 34 | ANKRD27 | 9054 | 0.331 | -0.2530 | No | ||
| 35 | OSBP | 9081 | 0.327 | -0.2525 | No | ||
| 36 | SPSB4 | 9278 | 0.284 | -0.2614 | No | ||
| 37 | RAB11FIP2 | 9572 | 0.217 | -0.2760 | No | ||
| 38 | ZIC1 | 9924 | 0.143 | -0.2941 | No | ||
| 39 | GJA1 | 10216 | 0.082 | -0.3094 | No | ||
| 40 | WBP2 | 10742 | -0.037 | -0.3375 | No | ||
| 41 | ENTPD5 | 11072 | -0.110 | -0.3546 | No | ||
| 42 | MAP4K4 | 11586 | -0.226 | -0.3810 | No | ||
| 43 | CHCHD4 | 11861 | -0.290 | -0.3941 | No | ||
| 44 | RCN1 | 12025 | -0.331 | -0.4010 | No | ||
| 45 | HIPK1 | 12159 | -0.362 | -0.4061 | No | ||
| 46 | KPNA3 | 12330 | -0.403 | -0.4129 | No | ||
| 47 | SLITRK2 | 12542 | -0.459 | -0.4216 | No | ||
| 48 | NRK | 12703 | -0.496 | -0.4274 | No | ||
| 49 | EBF3 | 12812 | -0.523 | -0.4301 | No | ||
| 50 | AUH | 13090 | -0.600 | -0.4416 | No | ||
| 51 | USP9X | 13424 | -0.697 | -0.4555 | No | ||
| 52 | CLASP1 | 13461 | -0.707 | -0.4533 | No | ||
| 53 | MRC1 | 13775 | -0.805 | -0.4655 | No | ||
| 54 | TBPL1 | 13815 | -0.815 | -0.4628 | No | ||
| 55 | KCNJ3 | 13821 | -0.816 | -0.4583 | No | ||
| 56 | UTX | 13836 | -0.819 | -0.4543 | No | ||
| 57 | NR4A3 | 13850 | -0.823 | -0.4502 | No | ||
| 58 | NFIX | 13920 | -0.840 | -0.4490 | No | ||
| 59 | ACAT2 | 13994 | -0.862 | -0.4478 | No | ||
| 60 | PPARGC1A | 14078 | -0.893 | -0.4471 | No | ||
| 61 | WTAP | 14106 | -0.905 | -0.4433 | No | ||
| 62 | RAB10 | 14942 | -1.183 | -0.4814 | No | ||
| 63 | STX12 | 15354 | -1.343 | -0.4958 | No | ||
| 64 | TMPO | 15636 | -1.460 | -0.5024 | No | ||
| 65 | RPS6KA3 | 15856 | -1.562 | -0.5051 | No | ||
| 66 | ULK2 | 16069 | -1.657 | -0.5069 | Yes | ||
| 67 | DDX3X | 16124 | -1.681 | -0.4999 | Yes | ||
| 68 | LRRC4 | 16131 | -1.684 | -0.4904 | Yes | ||
| 69 | HOOK3 | 16204 | -1.717 | -0.4842 | Yes | ||
| 70 | RSBN1 | 16360 | -1.791 | -0.4821 | Yes | ||
| 71 | ASF1A | 16393 | -1.808 | -0.4732 | Yes | ||
| 72 | PITPNA | 16474 | -1.842 | -0.4668 | Yes | ||
| 73 | CSNK1G3 | 16567 | -1.891 | -0.4607 | Yes | ||
| 74 | TNRC6A | 16743 | -1.984 | -0.4585 | Yes | ||
| 75 | UHRF2 | 16772 | -2.003 | -0.4483 | Yes | ||
| 76 | UBL3 | 16825 | -2.031 | -0.4392 | Yes | ||
| 77 | CUL1 | 16836 | -2.035 | -0.4278 | Yes | ||
| 78 | CLCN3 | 16985 | -2.113 | -0.4234 | Yes | ||
| 79 | MAPRE1 | 17095 | -2.168 | -0.4166 | Yes | ||
| 80 | SLMAP | 17147 | -2.204 | -0.4065 | Yes | ||
| 81 | ZDHHC21 | 17191 | -2.227 | -0.3957 | Yes | ||
| 82 | PJA2 | 17221 | -2.243 | -0.3842 | Yes | ||
| 83 | CEPT1 | 17222 | -2.244 | -0.3710 | Yes | ||
| 84 | FEM1B | 17259 | -2.264 | -0.3597 | Yes | ||
| 85 | SUMO1 | 17460 | -2.384 | -0.3565 | Yes | ||
| 86 | HS2ST1 | 17580 | -2.459 | -0.3486 | Yes | ||
| 87 | TRIM33 | 17582 | -2.459 | -0.3342 | Yes | ||
| 88 | ZNF318 | 17642 | -2.500 | -0.3228 | Yes | ||
| 89 | PPP1CB | 17671 | -2.520 | -0.3095 | Yes | ||
| 90 | ARFIP1 | 17696 | -2.534 | -0.2960 | Yes | ||
| 91 | DCUN1D1 | 17719 | -2.551 | -0.2822 | Yes | ||
| 92 | RANBP9 | 17783 | -2.593 | -0.2704 | Yes | ||
| 93 | DHX36 | 17848 | -2.642 | -0.2584 | Yes | ||
| 94 | KPNA1 | 17888 | -2.676 | -0.2448 | Yes | ||
| 95 | NDEL1 | 17946 | -2.716 | -0.2320 | Yes | ||
| 96 | ZHX1 | 18035 | -2.796 | -0.2204 | Yes | ||
| 97 | TOP1 | 18114 | -2.872 | -0.2078 | Yes | ||
| 98 | RAB1A | 18177 | -2.944 | -0.1939 | Yes | ||
| 99 | NFAT5 | 18208 | -2.982 | -0.1780 | Yes | ||
| 100 | TOP2B | 18210 | -2.984 | -0.1606 | Yes | ||
| 101 | PDCD10 | 18239 | -3.036 | -0.1443 | Yes | ||
| 102 | SMARCAD1 | 18290 | -3.118 | -0.1288 | Yes | ||
| 103 | REV3L | 18312 | -3.147 | -0.1115 | Yes | ||
| 104 | SFRS1 | 18333 | -3.183 | -0.0939 | Yes | ||
| 105 | SATB1 | 18414 | -3.343 | -0.0787 | Yes | ||
| 106 | ATP11B | 18483 | -3.537 | -0.0616 | Yes | ||
| 107 | POU4F2 | 18534 | -3.788 | -0.0421 | Yes | ||
| 108 | CCND1 | 18545 | -3.865 | -0.0200 | Yes | ||
| 109 | TGFA | 18567 | -4.065 | 0.0026 | Yes |
