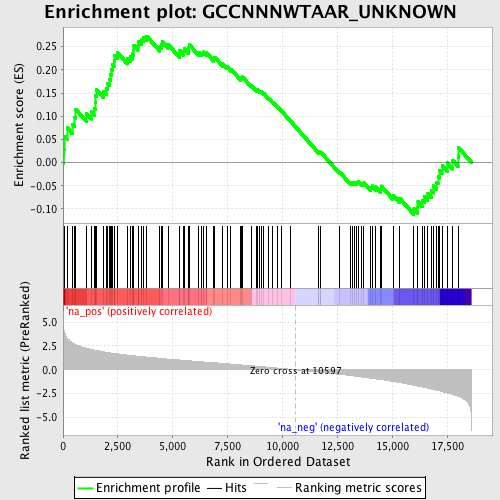
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GCCNNNWTAAR_UNKNOWN |
| Enrichment Score (ES) | 0.2726966 |
| Normalized Enrichment Score (NES) | 1.261484 |
| Nominal p-value | 0.10121457 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ETV6 | 40 | 4.153 | 0.0288 | Yes | ||
| 2 | GNAQ | 64 | 3.955 | 0.0570 | Yes | ||
| 3 | ETV5 | 182 | 3.305 | 0.0753 | Yes | ||
| 4 | CAMK2D | 432 | 2.810 | 0.0828 | Yes | ||
| 5 | VASP | 522 | 2.698 | 0.0980 | Yes | ||
| 6 | GGA1 | 567 | 2.648 | 0.1154 | Yes | ||
| 7 | HTR2C | 1064 | 2.244 | 0.1053 | Yes | ||
| 8 | CTDSP1 | 1277 | 2.121 | 0.1097 | Yes | ||
| 9 | LDHD | 1421 | 2.055 | 0.1172 | Yes | ||
| 10 | RPP21 | 1463 | 2.036 | 0.1302 | Yes | ||
| 11 | RPL10 | 1490 | 2.030 | 0.1439 | Yes | ||
| 12 | FYN | 1528 | 2.012 | 0.1569 | Yes | ||
| 13 | PEX5 | 1845 | 1.887 | 0.1539 | Yes | ||
| 14 | LRP5 | 1972 | 1.834 | 0.1607 | Yes | ||
| 15 | CDKN2C | 2038 | 1.811 | 0.1707 | Yes | ||
| 16 | SLC25A10 | 2130 | 1.775 | 0.1790 | Yes | ||
| 17 | GRASP | 2174 | 1.760 | 0.1898 | Yes | ||
| 18 | EGR1 | 2211 | 1.746 | 0.2008 | Yes | ||
| 19 | HOXB8 | 2246 | 1.734 | 0.2119 | Yes | ||
| 20 | BRUNOL4 | 2324 | 1.707 | 0.2205 | Yes | ||
| 21 | ATP5G2 | 2357 | 1.698 | 0.2314 | Yes | ||
| 22 | DAB1 | 2489 | 1.663 | 0.2367 | Yes | ||
| 23 | INPPL1 | 2937 | 1.538 | 0.2240 | Yes | ||
| 24 | CCBL1 | 3065 | 1.504 | 0.2284 | Yes | ||
| 25 | FES | 3160 | 1.479 | 0.2343 | Yes | ||
| 26 | HOXA10 | 3222 | 1.462 | 0.2419 | Yes | ||
| 27 | SOX5 | 3225 | 1.461 | 0.2527 | Yes | ||
| 28 | SLC39A5 | 3414 | 1.416 | 0.2531 | Yes | ||
| 29 | NKX2-5 | 3448 | 1.408 | 0.2618 | Yes | ||
| 30 | FHL3 | 3589 | 1.378 | 0.2645 | Yes | ||
| 31 | MMP24 | 3668 | 1.359 | 0.2704 | Yes | ||
| 32 | KCTD15 | 3809 | 1.328 | 0.2727 | Yes | ||
| 33 | PTPRG | 4408 | 1.198 | 0.2493 | No | ||
| 34 | CDKN1B | 4465 | 1.185 | 0.2551 | No | ||
| 35 | RAB33A | 4511 | 1.175 | 0.2614 | No | ||
| 36 | PDGFB | 4781 | 1.119 | 0.2553 | No | ||
| 37 | COCH | 5307 | 1.021 | 0.2345 | No | ||
| 38 | TRIB2 | 5321 | 1.019 | 0.2414 | No | ||
| 39 | VAX1 | 5505 | 0.981 | 0.2388 | No | ||
| 40 | NDRG2 | 5515 | 0.980 | 0.2456 | No | ||
| 41 | ATOH1 | 5720 | 0.944 | 0.2416 | No | ||
| 42 | FGF9 | 5732 | 0.942 | 0.2481 | No | ||
| 43 | BCL9 | 5741 | 0.940 | 0.2546 | No | ||
| 44 | H2AFX | 6170 | 0.860 | 0.2379 | No | ||
| 45 | ABCA1 | 6317 | 0.834 | 0.2363 | No | ||
| 46 | GDNF | 6389 | 0.822 | 0.2385 | No | ||
| 47 | HOXA4 | 6537 | 0.795 | 0.2365 | No | ||
| 48 | EGFL6 | 6868 | 0.732 | 0.2242 | No | ||
| 49 | MEF2D | 6907 | 0.727 | 0.2275 | No | ||
| 50 | DPF3 | 7272 | 0.667 | 0.2128 | No | ||
| 51 | JPH4 | 7470 | 0.631 | 0.2069 | No | ||
| 52 | PHOX2B | 7649 | 0.597 | 0.2017 | No | ||
| 53 | RFX4 | 8101 | 0.511 | 0.1812 | No | ||
| 54 | ELAVL4 | 8121 | 0.507 | 0.1839 | No | ||
| 55 | NPTX1 | 8175 | 0.496 | 0.1848 | No | ||
| 56 | MMP9 | 8581 | 0.419 | 0.1660 | No | ||
| 57 | SFTPC | 8834 | 0.376 | 0.1552 | No | ||
| 58 | STK35 | 8850 | 0.373 | 0.1572 | No | ||
| 59 | IMPDH2 | 8970 | 0.350 | 0.1533 | No | ||
| 60 | NFATC4 | 9023 | 0.339 | 0.1531 | No | ||
| 61 | NFIL3 | 9133 | 0.315 | 0.1495 | No | ||
| 62 | TBCC | 9375 | 0.263 | 0.1385 | No | ||
| 63 | BHLHB3 | 9556 | 0.221 | 0.1304 | No | ||
| 64 | ISL1 | 9792 | 0.168 | 0.1189 | No | ||
| 65 | MSX1 | 9947 | 0.138 | 0.1117 | No | ||
| 66 | ADAMTS5 | 10373 | 0.048 | 0.0891 | No | ||
| 67 | SHH | 11650 | -0.239 | 0.0219 | No | ||
| 68 | KCNH3 | 11654 | -0.240 | 0.0236 | No | ||
| 69 | SLC5A7 | 11723 | -0.257 | 0.0218 | No | ||
| 70 | NPR3 | 11744 | -0.262 | 0.0227 | No | ||
| 71 | SFXN5 | 12606 | -0.475 | -0.0203 | No | ||
| 72 | VCL | 13115 | -0.607 | -0.0432 | No | ||
| 73 | ASGR1 | 13202 | -0.629 | -0.0432 | No | ||
| 74 | ADAM12 | 13283 | -0.655 | -0.0426 | No | ||
| 75 | MITF | 13381 | -0.686 | -0.0428 | No | ||
| 76 | TGFB3 | 13443 | -0.702 | -0.0408 | No | ||
| 77 | APBA2BP | 13620 | -0.758 | -0.0447 | No | ||
| 78 | SOCS2 | 13712 | -0.784 | -0.0438 | No | ||
| 79 | OTP | 14027 | -0.876 | -0.0542 | No | ||
| 80 | PPARGC1A | 14078 | -0.893 | -0.0502 | No | ||
| 81 | ABCC5 | 14247 | -0.948 | -0.0523 | No | ||
| 82 | KLC2 | 14477 | -1.020 | -0.0570 | No | ||
| 83 | FOXP2 | 14497 | -1.024 | -0.0504 | No | ||
| 84 | HMGA2 | 15036 | -1.219 | -0.0704 | No | ||
| 85 | EML4 | 15341 | -1.339 | -0.0769 | No | ||
| 86 | SOX4 | 15990 | -1.618 | -0.0998 | No | ||
| 87 | XPO7 | 16158 | -1.696 | -0.0962 | No | ||
| 88 | MID1 | 16173 | -1.701 | -0.0843 | No | ||
| 89 | ITGA3 | 16364 | -1.792 | -0.0812 | No | ||
| 90 | EN2 | 16472 | -1.841 | -0.0733 | No | ||
| 91 | OTX1 | 16622 | -1.922 | -0.0670 | No | ||
| 92 | MPP2 | 16779 | -2.007 | -0.0605 | No | ||
| 93 | ZADH2 | 16874 | -2.055 | -0.0503 | No | ||
| 94 | RBMX | 17023 | -2.135 | -0.0423 | No | ||
| 95 | UBE2D3 | 17090 | -2.167 | -0.0298 | No | ||
| 96 | ARID1A | 17156 | -2.208 | -0.0168 | No | ||
| 97 | ITPR1 | 17270 | -2.271 | -0.0060 | No | ||
| 98 | PHEX | 17502 | -2.412 | -0.0005 | No | ||
| 99 | PTMA | 17749 | -2.571 | 0.0053 | No | ||
| 100 | HPS3 | 18003 | -2.770 | 0.0123 | No | ||
| 101 | ERG | 18027 | -2.789 | 0.0318 | No |
