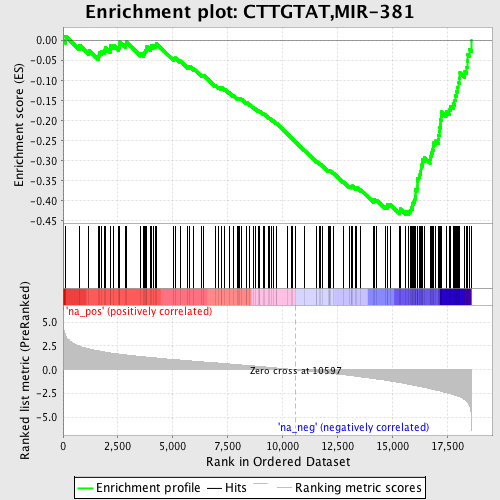
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CTTGTAT,MIR-381 |
| Enrichment Score (ES) | -0.43427613 |
| Normalized Enrichment Score (NES) | -2.1255035 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.5570483E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DLC1 | 109 | 3.570 | 0.0115 | No | ||
| 2 | BAT2 | 749 | 2.469 | -0.0111 | No | ||
| 3 | GFRA1 | 1178 | 2.177 | -0.0237 | No | ||
| 4 | CACNA1C | 1624 | 1.973 | -0.0381 | No | ||
| 5 | SEMA6D | 1641 | 1.966 | -0.0294 | No | ||
| 6 | RNF13 | 1757 | 1.920 | -0.0263 | No | ||
| 7 | KLF12 | 1869 | 1.873 | -0.0232 | No | ||
| 8 | DHX40 | 1933 | 1.848 | -0.0176 | No | ||
| 9 | RPS6KB1 | 2143 | 1.770 | -0.0203 | No | ||
| 10 | LPHN1 | 2164 | 1.765 | -0.0128 | No | ||
| 11 | TRIM63 | 2308 | 1.713 | -0.0122 | No | ||
| 12 | ANKRD50 | 2531 | 1.654 | -0.0161 | No | ||
| 13 | FLRT3 | 2579 | 1.642 | -0.0107 | No | ||
| 14 | GAD1 | 2588 | 1.639 | -0.0031 | No | ||
| 15 | ELOVL7 | 2842 | 1.567 | -0.0092 | No | ||
| 16 | SRRM1 | 2869 | 1.556 | -0.0030 | No | ||
| 17 | CLCF1 | 3521 | 1.393 | -0.0315 | No | ||
| 18 | HNRPR | 3667 | 1.360 | -0.0327 | No | ||
| 19 | JPH1 | 3722 | 1.346 | -0.0291 | No | ||
| 20 | PMP22 | 3751 | 1.339 | -0.0241 | No | ||
| 21 | KCTD15 | 3809 | 1.328 | -0.0207 | No | ||
| 22 | ETS1 | 3817 | 1.325 | -0.0146 | No | ||
| 23 | COL3A1 | 3999 | 1.288 | -0.0182 | No | ||
| 24 | UBE2E2 | 4008 | 1.287 | -0.0123 | No | ||
| 25 | PNRC1 | 4122 | 1.263 | -0.0123 | No | ||
| 26 | TBC1D15 | 4228 | 1.240 | -0.0119 | No | ||
| 27 | EPHA3 | 4246 | 1.237 | -0.0068 | No | ||
| 28 | MTSS1 | 5028 | 1.076 | -0.0439 | No | ||
| 29 | AGPAT3 | 5107 | 1.060 | -0.0430 | No | ||
| 30 | TMEM16D | 5340 | 1.015 | -0.0506 | No | ||
| 31 | PDGFC | 5681 | 0.951 | -0.0643 | No | ||
| 32 | ZFPM2 | 5771 | 0.934 | -0.0646 | No | ||
| 33 | NFKBIA | 5957 | 0.899 | -0.0703 | No | ||
| 34 | ABCA1 | 6317 | 0.834 | -0.0856 | No | ||
| 35 | ZCCHC11 | 6410 | 0.817 | -0.0866 | No | ||
| 36 | HBP1 | 6927 | 0.724 | -0.1110 | No | ||
| 37 | SLC24A3 | 7098 | 0.699 | -0.1168 | No | ||
| 38 | FOXF2 | 7204 | 0.679 | -0.1192 | No | ||
| 39 | WNT5A | 7213 | 0.677 | -0.1164 | No | ||
| 40 | BIRC6 | 7356 | 0.654 | -0.1209 | No | ||
| 41 | RNF144 | 7573 | 0.612 | -0.1296 | No | ||
| 42 | RNF2 | 7751 | 0.575 | -0.1364 | No | ||
| 43 | CEBPA | 7963 | 0.537 | -0.1452 | No | ||
| 44 | ACVR2A | 7995 | 0.531 | -0.1443 | No | ||
| 45 | SEMA3C | 8031 | 0.525 | -0.1436 | No | ||
| 46 | NAP1L5 | 8117 | 0.508 | -0.1457 | No | ||
| 47 | OSBPL3 | 8344 | 0.466 | -0.1557 | No | ||
| 48 | BTBD7 | 8375 | 0.461 | -0.1551 | No | ||
| 49 | ACTG1 | 8503 | 0.434 | -0.1598 | No | ||
| 50 | NFIA | 8690 | 0.401 | -0.1680 | No | ||
| 51 | PARD6B | 8749 | 0.390 | -0.1692 | No | ||
| 52 | ACSL3 | 8893 | 0.365 | -0.1752 | No | ||
| 53 | GRM8 | 8950 | 0.353 | -0.1765 | No | ||
| 54 | ZFAND3 | 8971 | 0.350 | -0.1759 | No | ||
| 55 | ST8SIA2 | 9116 | 0.320 | -0.1821 | No | ||
| 56 | VSNL1 | 9169 | 0.305 | -0.1834 | No | ||
| 57 | SHANK2 | 9348 | 0.269 | -0.1918 | No | ||
| 58 | BCL11A | 9418 | 0.251 | -0.1943 | No | ||
| 59 | UBN1 | 9494 | 0.234 | -0.1972 | No | ||
| 60 | RAB11FIP2 | 9572 | 0.217 | -0.2003 | No | ||
| 61 | NR5A2 | 9707 | 0.187 | -0.2066 | No | ||
| 62 | B3GNT5 | 9710 | 0.186 | -0.2058 | No | ||
| 63 | GJA1 | 10216 | 0.082 | -0.2328 | No | ||
| 64 | RPL35A | 10386 | 0.045 | -0.2417 | No | ||
| 65 | PAPOLA | 10461 | 0.030 | -0.2456 | No | ||
| 66 | UBE4A | 10610 | -0.003 | -0.2536 | No | ||
| 67 | RHOQ | 11009 | -0.096 | -0.2747 | No | ||
| 68 | ARID4B | 11551 | -0.218 | -0.3029 | No | ||
| 69 | BHLHB5 | 11553 | -0.218 | -0.3019 | No | ||
| 70 | MDS1 | 11685 | -0.246 | -0.3078 | No | ||
| 71 | THRAP1 | 11739 | -0.262 | -0.3094 | No | ||
| 72 | EPHA4 | 11818 | -0.279 | -0.3122 | No | ||
| 73 | GRM1 | 12108 | -0.348 | -0.3262 | No | ||
| 74 | PHF15 | 12120 | -0.351 | -0.3251 | No | ||
| 75 | HIPK1 | 12159 | -0.362 | -0.3254 | No | ||
| 76 | SLC6A8 | 12196 | -0.369 | -0.3255 | No | ||
| 77 | KPNA3 | 12330 | -0.403 | -0.3308 | No | ||
| 78 | THRAP2 | 12787 | -0.516 | -0.3529 | No | ||
| 79 | CRSP2 | 13054 | -0.591 | -0.3645 | No | ||
| 80 | KIF1B | 13122 | -0.610 | -0.3651 | No | ||
| 81 | JAG2 | 13157 | -0.618 | -0.3640 | No | ||
| 82 | SSH2 | 13194 | -0.627 | -0.3629 | No | ||
| 83 | LZTS2 | 13346 | -0.677 | -0.3677 | No | ||
| 84 | MITF | 13381 | -0.686 | -0.3662 | No | ||
| 85 | R3HDM1 | 13572 | -0.740 | -0.3729 | No | ||
| 86 | CXCR4 | 14140 | -0.915 | -0.3992 | No | ||
| 87 | GORASP2 | 14185 | -0.928 | -0.3970 | No | ||
| 88 | KCTD3 | 14293 | -0.962 | -0.3981 | No | ||
| 89 | PDAP1 | 14704 | -1.101 | -0.4150 | No | ||
| 90 | PBEF1 | 14773 | -1.126 | -0.4132 | No | ||
| 91 | WEE1 | 14784 | -1.131 | -0.4082 | No | ||
| 92 | HNRPH2 | 14916 | -1.174 | -0.4096 | No | ||
| 93 | CGGBP1 | 15351 | -1.342 | -0.4265 | No | ||
| 94 | STX12 | 15354 | -1.343 | -0.4201 | No | ||
| 95 | PHF2 | 15610 | -1.449 | -0.4269 | No | ||
| 96 | GAS2 | 15748 | -1.512 | -0.4269 | Yes | ||
| 97 | QKI | 15817 | -1.540 | -0.4231 | Yes | ||
| 98 | RPS6KA3 | 15856 | -1.562 | -0.4175 | Yes | ||
| 99 | SIPA1L2 | 15924 | -1.588 | -0.4134 | Yes | ||
| 100 | ZMYND11 | 15939 | -1.594 | -0.4064 | Yes | ||
| 101 | SOX4 | 15990 | -1.618 | -0.4013 | Yes | ||
| 102 | INSIG1 | 16034 | -1.644 | -0.3956 | Yes | ||
| 103 | RBM16 | 16043 | -1.646 | -0.3880 | Yes | ||
| 104 | KLF4 | 16058 | -1.654 | -0.3807 | Yes | ||
| 105 | EIF1 | 16059 | -1.655 | -0.3726 | Yes | ||
| 106 | LRRC4 | 16131 | -1.684 | -0.3683 | Yes | ||
| 107 | SNRK | 16146 | -1.690 | -0.3608 | Yes | ||
| 108 | CCNT2 | 16150 | -1.693 | -0.3527 | Yes | ||
| 109 | XPO7 | 16158 | -1.696 | -0.3448 | Yes | ||
| 110 | SLC38A2 | 16220 | -1.723 | -0.3397 | Yes | ||
| 111 | UBE3A | 16263 | -1.740 | -0.3335 | Yes | ||
| 112 | DDX6 | 16290 | -1.756 | -0.3264 | Yes | ||
| 113 | AZIN1 | 16311 | -1.768 | -0.3189 | Yes | ||
| 114 | NIN | 16315 | -1.770 | -0.3104 | Yes | ||
| 115 | RALGPS1 | 16374 | -1.796 | -0.3048 | Yes | ||
| 116 | ITCH | 16395 | -1.809 | -0.2971 | Yes | ||
| 117 | ASXL2 | 16471 | -1.841 | -0.2922 | Yes | ||
| 118 | NRXN1 | 16724 | -1.977 | -0.2962 | Yes | ||
| 119 | BCL11B | 16759 | -1.998 | -0.2883 | Yes | ||
| 120 | KHDRBS1 | 16794 | -2.018 | -0.2803 | Yes | ||
| 121 | EIF4G2 | 16823 | -2.030 | -0.2719 | Yes | ||
| 122 | RNF111 | 16875 | -2.055 | -0.2647 | Yes | ||
| 123 | GTF2I | 16891 | -2.062 | -0.2555 | Yes | ||
| 124 | CHD4 | 16990 | -2.116 | -0.2505 | Yes | ||
| 125 | ZFYVE20 | 17099 | -2.170 | -0.2457 | Yes | ||
| 126 | APRIN | 17114 | -2.181 | -0.2359 | Yes | ||
| 127 | SLMAP | 17147 | -2.204 | -0.2269 | Yes | ||
| 128 | MBNL2 | 17164 | -2.211 | -0.2170 | Yes | ||
| 129 | RAP2C | 17200 | -2.232 | -0.2080 | Yes | ||
| 130 | ETF1 | 17211 | -2.239 | -0.1976 | Yes | ||
| 131 | ATRN | 17234 | -2.250 | -0.1879 | Yes | ||
| 132 | PSIP1 | 17253 | -2.263 | -0.1778 | Yes | ||
| 133 | SYNCRIP | 17454 | -2.383 | -0.1770 | Yes | ||
| 134 | PURG | 17601 | -2.478 | -0.1729 | Yes | ||
| 135 | EIF3S10 | 17651 | -2.505 | -0.1633 | Yes | ||
| 136 | YTHDF1 | 17776 | -2.586 | -0.1574 | Yes | ||
| 137 | EIF4G3 | 17856 | -2.652 | -0.1488 | Yes | ||
| 138 | NLK | 17875 | -2.665 | -0.1368 | Yes | ||
| 139 | UBR1 | 17929 | -2.703 | -0.1265 | Yes | ||
| 140 | NDUFAB1 | 17988 | -2.756 | -0.1162 | Yes | ||
| 141 | STAG1 | 17999 | -2.762 | -0.1033 | Yes | ||
| 142 | CNOT7 | 18067 | -2.828 | -0.0931 | Yes | ||
| 143 | CSTF3 | 18086 | -2.844 | -0.0803 | Yes | ||
| 144 | ROD1 | 18313 | -3.150 | -0.0772 | Yes | ||
| 145 | ROCK2 | 18399 | -3.300 | -0.0657 | Yes | ||
| 146 | SATB1 | 18414 | -3.343 | -0.0502 | Yes | ||
| 147 | PAPOLG | 18429 | -3.392 | -0.0344 | Yes | ||
| 148 | APPBP2 | 18510 | -3.674 | -0.0208 | Yes | ||
| 149 | BRD7 | 18613 | -5.446 | 0.0002 | Yes |
