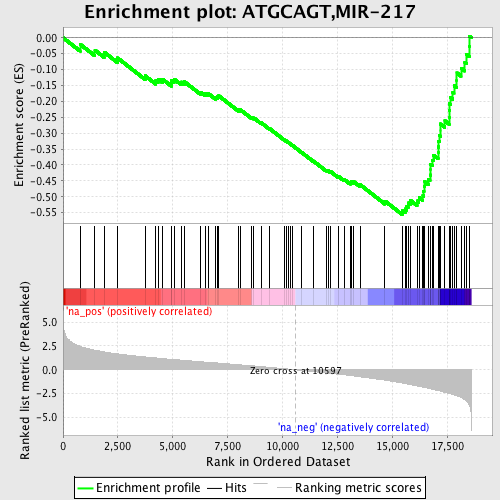
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | -0.55570954 |
| Normalized Enrichment Score (NES) | -2.4500172 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TACC2 | 774 | 2.456 | -0.0203 | No | ||
| 2 | FBN2 | 1452 | 2.042 | -0.0389 | No | ||
| 3 | KLF12 | 1869 | 1.873 | -0.0450 | No | ||
| 4 | CAPN3 | 2479 | 1.666 | -0.0633 | No | ||
| 5 | SEPT4 | 3741 | 1.342 | -0.1196 | No | ||
| 6 | TBC1D15 | 4228 | 1.240 | -0.1350 | No | ||
| 7 | NR4A2 | 4328 | 1.217 | -0.1297 | No | ||
| 8 | CHN2 | 4540 | 1.167 | -0.1308 | No | ||
| 9 | KCNA3 | 4932 | 1.092 | -0.1424 | No | ||
| 10 | STAT5B | 4945 | 1.090 | -0.1335 | No | ||
| 11 | POLG | 5057 | 1.071 | -0.1301 | No | ||
| 12 | A2BP1 | 5385 | 1.005 | -0.1390 | No | ||
| 13 | CUL5 | 5535 | 0.976 | -0.1385 | No | ||
| 14 | GPM6A | 6271 | 0.842 | -0.1708 | No | ||
| 15 | DLGAP2 | 6473 | 0.805 | -0.1746 | No | ||
| 16 | STRBP | 6614 | 0.782 | -0.1753 | No | ||
| 17 | NEUROD1 | 6959 | 0.720 | -0.1876 | No | ||
| 18 | PAQR3 | 7039 | 0.708 | -0.1856 | No | ||
| 19 | BAI3 | 7099 | 0.699 | -0.1827 | No | ||
| 20 | ACVR2A | 7995 | 0.531 | -0.2263 | No | ||
| 21 | ST18 | 8082 | 0.515 | -0.2265 | No | ||
| 22 | TMSB4X | 8596 | 0.417 | -0.2505 | No | ||
| 23 | NFIA | 8690 | 0.401 | -0.2520 | No | ||
| 24 | MAMDC2 | 9029 | 0.338 | -0.2673 | No | ||
| 25 | BCL11A | 9418 | 0.251 | -0.2860 | No | ||
| 26 | STX6 | 10106 | 0.108 | -0.3221 | No | ||
| 27 | DMRT2 | 10161 | 0.094 | -0.3242 | No | ||
| 28 | ZCCHC2 | 10282 | 0.069 | -0.3301 | No | ||
| 29 | ADAMTS5 | 10373 | 0.048 | -0.3345 | No | ||
| 30 | RIN2 | 10451 | 0.031 | -0.3384 | No | ||
| 31 | FBXW11 | 10879 | -0.066 | -0.3609 | No | ||
| 32 | SOX11 | 11402 | -0.185 | -0.3874 | No | ||
| 33 | GDI2 | 11992 | -0.322 | -0.4164 | No | ||
| 34 | GRIK2 | 12074 | -0.341 | -0.4177 | No | ||
| 35 | DACH1 | 12169 | -0.364 | -0.4196 | No | ||
| 36 | PSMF1 | 12552 | -0.462 | -0.4362 | No | ||
| 37 | JOSD3 | 12811 | -0.523 | -0.4455 | No | ||
| 38 | FN1 | 13081 | -0.597 | -0.4548 | No | ||
| 39 | STX1A | 13130 | -0.611 | -0.4521 | No | ||
| 40 | DYRK1A | 13236 | -0.637 | -0.4522 | No | ||
| 41 | AKAP2 | 13542 | -0.730 | -0.4622 | No | ||
| 42 | ELOVL4 | 14669 | -1.086 | -0.5135 | No | ||
| 43 | YAF2 | 15453 | -1.388 | -0.5436 | Yes | ||
| 44 | HIVEP3 | 15590 | -1.443 | -0.5383 | Yes | ||
| 45 | SENP5 | 15665 | -1.475 | -0.5294 | Yes | ||
| 46 | RTF1 | 15725 | -1.499 | -0.5195 | Yes | ||
| 47 | ATP11C | 15819 | -1.541 | -0.5110 | Yes | ||
| 48 | IVNS1ABP | 16132 | -1.685 | -0.5131 | Yes | ||
| 49 | SLC38A2 | 16220 | -1.723 | -0.5027 | Yes | ||
| 50 | ATP1B1 | 16399 | -1.812 | -0.4965 | Yes | ||
| 51 | STAG2 | 16433 | -1.825 | -0.4823 | Yes | ||
| 52 | EZH2 | 16450 | -1.831 | -0.4672 | Yes | ||
| 53 | DYNC1LI2 | 16486 | -1.848 | -0.4529 | Yes | ||
| 54 | HNRPUL1 | 16657 | -1.935 | -0.4451 | Yes | ||
| 55 | MSN | 16723 | -1.970 | -0.4314 | Yes | ||
| 56 | YWHAG | 16732 | -1.980 | -0.4145 | Yes | ||
| 57 | ATP6V1A | 16736 | -1.981 | -0.3974 | Yes | ||
| 58 | UBL3 | 16825 | -2.031 | -0.3844 | Yes | ||
| 59 | SIRT1 | 16877 | -2.056 | -0.3692 | Yes | ||
| 60 | PCNA | 17091 | -2.167 | -0.3617 | Yes | ||
| 61 | ZFYVE20 | 17099 | -2.170 | -0.3431 | Yes | ||
| 62 | APRIN | 17114 | -2.181 | -0.3248 | Yes | ||
| 63 | MAPK1 | 17150 | -2.206 | -0.3074 | Yes | ||
| 64 | KCTD9 | 17199 | -2.230 | -0.2905 | Yes | ||
| 65 | RAP2C | 17200 | -2.232 | -0.2709 | Yes | ||
| 66 | KRAS | 17403 | -2.349 | -0.2613 | Yes | ||
| 67 | EIF4A2 | 17593 | -2.470 | -0.2499 | Yes | ||
| 68 | MTPN | 17600 | -2.477 | -0.2286 | Yes | ||
| 69 | MYEF2 | 17611 | -2.483 | -0.2074 | Yes | ||
| 70 | SLC39A10 | 17667 | -2.516 | -0.1884 | Yes | ||
| 71 | SLC31A1 | 17763 | -2.577 | -0.1710 | Yes | ||
| 72 | TLK2 | 17816 | -2.616 | -0.1509 | Yes | ||
| 73 | NDEL1 | 17946 | -2.716 | -0.1341 | Yes | ||
| 74 | OGT | 17951 | -2.718 | -0.1106 | Yes | ||
| 75 | SLITRK3 | 18135 | -2.904 | -0.0950 | Yes | ||
| 76 | PPM1D | 18311 | -3.147 | -0.0770 | Yes | ||
| 77 | HNRPA2B1 | 18395 | -3.287 | -0.0527 | Yes | ||
| 78 | APPBP2 | 18510 | -3.674 | -0.0267 | Yes | ||
| 79 | EHMT1 | 18518 | -3.704 | 0.0053 | Yes |
