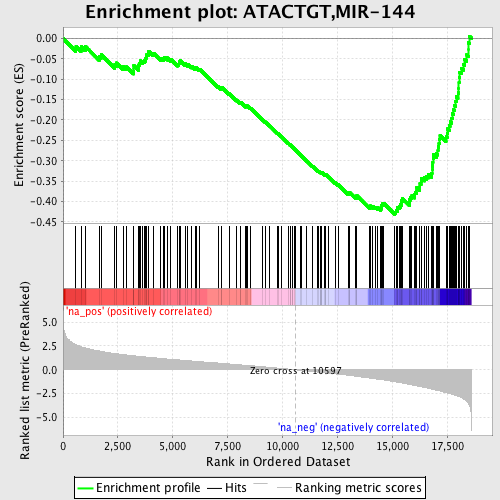
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATACTGT,MIR-144 |
| Enrichment Score (ES) | -0.43175194 |
| Normalized Enrichment Score (NES) | -2.1363554 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.5101527E-4 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DLG5 | 586 | 2.628 | -0.0190 | No | ||
| 2 | DGCR2 | 818 | 2.423 | -0.0198 | No | ||
| 3 | PTHLH | 1011 | 2.275 | -0.0192 | No | ||
| 4 | SEMA6D | 1641 | 1.966 | -0.0438 | No | ||
| 5 | FMR1 | 1729 | 1.931 | -0.0391 | No | ||
| 6 | ATP5G2 | 2357 | 1.698 | -0.0649 | No | ||
| 7 | PDE7B | 2419 | 1.681 | -0.0600 | No | ||
| 8 | TRIB1 | 2735 | 1.594 | -0.0694 | No | ||
| 9 | ATXN1 | 2880 | 1.552 | -0.0697 | No | ||
| 10 | SMPD3 | 3205 | 1.467 | -0.0801 | No | ||
| 11 | HOXA10 | 3222 | 1.462 | -0.0739 | No | ||
| 12 | PIM1 | 3224 | 1.461 | -0.0669 | No | ||
| 13 | CALCRL | 3436 | 1.410 | -0.0715 | No | ||
| 14 | PAPPA | 3444 | 1.409 | -0.0651 | No | ||
| 15 | NR2F2 | 3482 | 1.401 | -0.0603 | No | ||
| 16 | CPEB3 | 3508 | 1.397 | -0.0549 | No | ||
| 17 | WDFY3 | 3640 | 1.369 | -0.0554 | No | ||
| 18 | JPH1 | 3722 | 1.346 | -0.0532 | No | ||
| 19 | LHX2 | 3769 | 1.335 | -0.0493 | No | ||
| 20 | AGRN | 3785 | 1.331 | -0.0437 | No | ||
| 21 | ETS1 | 3817 | 1.325 | -0.0389 | No | ||
| 22 | DMD | 3883 | 1.309 | -0.0361 | No | ||
| 23 | MYST3 | 3903 | 1.304 | -0.0308 | No | ||
| 24 | PNRC1 | 4122 | 1.263 | -0.0365 | No | ||
| 25 | HDHD2 | 4450 | 1.189 | -0.0485 | No | ||
| 26 | CPEB2 | 4567 | 1.163 | -0.0491 | No | ||
| 27 | PTGFRN | 4606 | 1.154 | -0.0456 | No | ||
| 28 | SNAP91 | 4745 | 1.126 | -0.0476 | No | ||
| 29 | ALDH1A3 | 4905 | 1.097 | -0.0509 | No | ||
| 30 | NEDD4 | 5216 | 1.039 | -0.0627 | No | ||
| 31 | EDAR | 5281 | 1.027 | -0.0612 | No | ||
| 32 | TTN | 5301 | 1.022 | -0.0573 | No | ||
| 33 | STARD8 | 5341 | 1.015 | -0.0545 | No | ||
| 34 | CXCL12 | 5563 | 0.972 | -0.0618 | No | ||
| 35 | MEIS1 | 5679 | 0.952 | -0.0634 | No | ||
| 36 | ARID5B | 5848 | 0.920 | -0.0680 | No | ||
| 37 | ANK2 | 6021 | 0.888 | -0.0731 | No | ||
| 38 | RARB | 6070 | 0.879 | -0.0714 | No | ||
| 39 | BACH2 | 6215 | 0.851 | -0.0751 | No | ||
| 40 | GCNT2 | 7071 | 0.702 | -0.1180 | No | ||
| 41 | SEMA6A | 7211 | 0.678 | -0.1222 | No | ||
| 42 | CD28 | 7225 | 0.675 | -0.1197 | No | ||
| 43 | NRP2 | 7576 | 0.612 | -0.1357 | No | ||
| 44 | SFRP1 | 7906 | 0.547 | -0.1509 | No | ||
| 45 | ST18 | 8082 | 0.515 | -0.1578 | No | ||
| 46 | RFX4 | 8101 | 0.511 | -0.1563 | No | ||
| 47 | ST6GALNAC3 | 8325 | 0.469 | -0.1662 | No | ||
| 48 | MYO1E | 8342 | 0.466 | -0.1648 | No | ||
| 49 | TBX1 | 8395 | 0.458 | -0.1654 | No | ||
| 50 | LGR4 | 8531 | 0.430 | -0.1706 | No | ||
| 51 | SLITRK4 | 9089 | 0.325 | -0.1992 | No | ||
| 52 | IDH2 | 9218 | 0.294 | -0.2047 | No | ||
| 53 | WIF1 | 9407 | 0.254 | -0.2137 | No | ||
| 54 | FMN2 | 9768 | 0.174 | -0.2323 | No | ||
| 55 | ESRRG | 9810 | 0.163 | -0.2338 | No | ||
| 56 | MSX1 | 9947 | 0.138 | -0.2405 | No | ||
| 57 | ZCCHC2 | 10282 | 0.069 | -0.2582 | No | ||
| 58 | PPARA | 10343 | 0.053 | -0.2612 | No | ||
| 59 | SMARCA1 | 10344 | 0.053 | -0.2609 | No | ||
| 60 | RIN2 | 10451 | 0.031 | -0.2665 | No | ||
| 61 | ELL2 | 10453 | 0.031 | -0.2664 | No | ||
| 62 | SP4 | 10540 | 0.012 | -0.2710 | No | ||
| 63 | UBE4A | 10610 | -0.003 | -0.2748 | No | ||
| 64 | RGS16 | 10822 | -0.055 | -0.2859 | No | ||
| 65 | FBXW11 | 10879 | -0.066 | -0.2886 | No | ||
| 66 | HNRPF | 11111 | -0.115 | -0.3006 | No | ||
| 67 | CAV2 | 11362 | -0.175 | -0.3133 | No | ||
| 68 | LIFR | 11591 | -0.226 | -0.3245 | No | ||
| 69 | PAX3 | 11631 | -0.235 | -0.3255 | No | ||
| 70 | MYT1 | 11716 | -0.254 | -0.3288 | No | ||
| 71 | PANK1 | 11734 | -0.261 | -0.3285 | No | ||
| 72 | PCDH18 | 11777 | -0.270 | -0.3295 | No | ||
| 73 | PTPN9 | 11794 | -0.273 | -0.3290 | No | ||
| 74 | BRPF1 | 11930 | -0.306 | -0.3348 | No | ||
| 75 | CCT2 | 11951 | -0.312 | -0.3344 | No | ||
| 76 | FAM60A | 11961 | -0.315 | -0.3334 | No | ||
| 77 | AHCY | 12113 | -0.349 | -0.3399 | No | ||
| 78 | VKORC1L1 | 12412 | -0.422 | -0.3540 | No | ||
| 79 | CBX1 | 12541 | -0.458 | -0.3587 | No | ||
| 80 | NGFRAP1 | 13004 | -0.576 | -0.3809 | No | ||
| 81 | CRSP2 | 13054 | -0.591 | -0.3807 | No | ||
| 82 | FOXO1A | 13058 | -0.592 | -0.3780 | No | ||
| 83 | PLAG1 | 13330 | -0.670 | -0.3894 | No | ||
| 84 | SLC12A2 | 13380 | -0.685 | -0.3888 | No | ||
| 85 | FOSB | 13389 | -0.687 | -0.3859 | No | ||
| 86 | FBXL3 | 13986 | -0.860 | -0.4140 | No | ||
| 87 | PURA | 14002 | -0.866 | -0.4106 | No | ||
| 88 | WTAP | 14106 | -0.905 | -0.4118 | No | ||
| 89 | SLC7A11 | 14220 | -0.939 | -0.4134 | No | ||
| 90 | EIF5 | 14339 | -0.978 | -0.4151 | No | ||
| 91 | GATA3 | 14460 | -1.016 | -0.4167 | No | ||
| 92 | SORCS3 | 14495 | -1.023 | -0.4135 | No | ||
| 93 | PALM2 | 14517 | -1.031 | -0.4097 | No | ||
| 94 | PTP4A1 | 14534 | -1.036 | -0.4055 | No | ||
| 95 | CBX4 | 14615 | -1.065 | -0.4047 | No | ||
| 96 | FBXO32 | 15115 | -1.255 | -0.4257 | Yes | ||
| 97 | NR3C1 | 15172 | -1.276 | -0.4225 | Yes | ||
| 98 | ALS2 | 15229 | -1.300 | -0.4193 | Yes | ||
| 99 | GDF10 | 15247 | -1.308 | -0.4139 | Yes | ||
| 100 | FUSIP1 | 15328 | -1.334 | -0.4118 | Yes | ||
| 101 | PPP1R16B | 15374 | -1.351 | -0.4077 | Yes | ||
| 102 | CAPZA1 | 15415 | -1.370 | -0.4032 | Yes | ||
| 103 | CDH11 | 15436 | -1.378 | -0.3976 | Yes | ||
| 104 | RHOBTB1 | 15475 | -1.396 | -0.3929 | Yes | ||
| 105 | PUM1 | 15776 | -1.521 | -0.4018 | Yes | ||
| 106 | ARFGEF1 | 15799 | -1.532 | -0.3956 | Yes | ||
| 107 | QKI | 15817 | -1.540 | -0.3891 | Yes | ||
| 108 | ACBD3 | 15898 | -1.579 | -0.3858 | Yes | ||
| 109 | GSPT1 | 16003 | -1.629 | -0.3835 | Yes | ||
| 110 | TFRC | 16038 | -1.645 | -0.3774 | Yes | ||
| 111 | ATP2B2 | 16115 | -1.679 | -0.3734 | Yes | ||
| 112 | ARRDC3 | 16120 | -1.680 | -0.3655 | Yes | ||
| 113 | PHTF2 | 16255 | -1.734 | -0.3643 | Yes | ||
| 114 | FNDC3A | 16264 | -1.741 | -0.3563 | Yes | ||
| 115 | NIN | 16315 | -1.770 | -0.3505 | Yes | ||
| 116 | ZIC4 | 16338 | -1.782 | -0.3431 | Yes | ||
| 117 | SUCLA2 | 16461 | -1.837 | -0.3408 | Yes | ||
| 118 | CCNK | 16572 | -1.893 | -0.3376 | Yes | ||
| 119 | UBE2D2 | 16660 | -1.936 | -0.3329 | Yes | ||
| 120 | PKNOX1 | 16799 | -2.020 | -0.3306 | Yes | ||
| 121 | PDE4D | 16816 | -2.027 | -0.3217 | Yes | ||
| 122 | EIF4G2 | 16823 | -2.030 | -0.3122 | Yes | ||
| 123 | CCNE2 | 16850 | -2.041 | -0.3037 | Yes | ||
| 124 | MAPK6 | 16859 | -2.048 | -0.2942 | Yes | ||
| 125 | RNF111 | 16875 | -2.055 | -0.2851 | Yes | ||
| 126 | PABPN1 | 17007 | -2.123 | -0.2819 | Yes | ||
| 127 | PAFAH1B1 | 17084 | -2.165 | -0.2756 | Yes | ||
| 128 | UBE2D3 | 17090 | -2.167 | -0.2654 | Yes | ||
| 129 | SON | 17130 | -2.195 | -0.2569 | Yes | ||
| 130 | ARID1A | 17156 | -2.208 | -0.2475 | Yes | ||
| 131 | RBM12 | 17176 | -2.216 | -0.2378 | Yes | ||
| 132 | HAT1 | 17453 | -2.382 | -0.2413 | Yes | ||
| 133 | PFKFB2 | 17503 | -2.412 | -0.2322 | Yes | ||
| 134 | ZFX | 17528 | -2.430 | -0.2218 | Yes | ||
| 135 | MTPN | 17600 | -2.477 | -0.2136 | Yes | ||
| 136 | SLC23A2 | 17636 | -2.496 | -0.2035 | Yes | ||
| 137 | NFE2L2 | 17716 | -2.550 | -0.1954 | Yes | ||
| 138 | HNRPU | 17736 | -2.564 | -0.1840 | Yes | ||
| 139 | ACSL4 | 17802 | -2.604 | -0.1749 | Yes | ||
| 140 | CDH2 | 17829 | -2.624 | -0.1637 | Yes | ||
| 141 | KPNA1 | 17888 | -2.676 | -0.1539 | Yes | ||
| 142 | CUGBP2 | 17921 | -2.698 | -0.1425 | Yes | ||
| 143 | STAG1 | 17999 | -2.762 | -0.1333 | Yes | ||
| 144 | MARCKS | 18036 | -2.799 | -0.1218 | Yes | ||
| 145 | ITSN2 | 18042 | -2.802 | -0.1085 | Yes | ||
| 146 | SIN3A | 18050 | -2.813 | -0.0952 | Yes | ||
| 147 | GLCCI1 | 18078 | -2.834 | -0.0830 | Yes | ||
| 148 | ARID2 | 18148 | -2.918 | -0.0726 | Yes | ||
| 149 | NARG1 | 18243 | -3.040 | -0.0630 | Yes | ||
| 150 | CLK1 | 18297 | -3.127 | -0.0507 | Yes | ||
| 151 | HSF2 | 18400 | -3.301 | -0.0403 | Yes | ||
| 152 | PPP3R1 | 18473 | -3.514 | -0.0272 | Yes | ||
| 153 | SS18 | 18476 | -3.521 | -0.0102 | Yes | ||
| 154 | APPBP2 | 18510 | -3.674 | 0.0057 | Yes |
