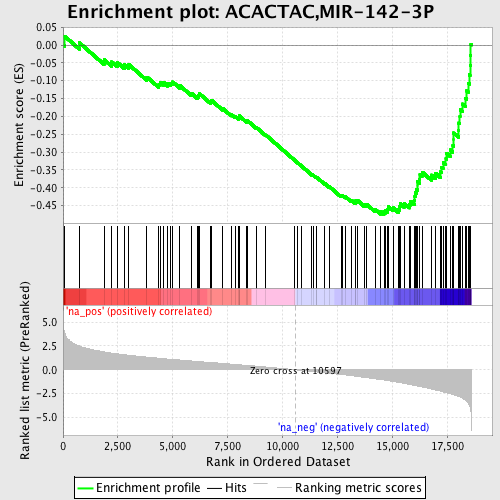
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ACACTAC,MIR-142-3P |
| Enrichment Score (ES) | -0.474944 |
| Normalized Enrichment Score (NES) | -2.1828701 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GNAQ | 64 | 3.955 | 0.0250 | No | ||
| 2 | COPS7A | 741 | 2.476 | 0.0063 | No | ||
| 3 | SP8 | 1868 | 1.873 | -0.0410 | No | ||
| 4 | CRK | 2219 | 1.743 | -0.0473 | No | ||
| 5 | AKT1S1 | 2475 | 1.667 | -0.0491 | No | ||
| 6 | ANK3 | 2794 | 1.579 | -0.0549 | No | ||
| 7 | PRLR | 2996 | 1.522 | -0.0548 | No | ||
| 8 | EDG3 | 3822 | 1.324 | -0.0898 | No | ||
| 9 | MAP3K11 | 4368 | 1.207 | -0.1105 | No | ||
| 10 | FKBP1A | 4419 | 1.195 | -0.1046 | No | ||
| 11 | CPEB2 | 4567 | 1.163 | -0.1042 | No | ||
| 12 | ARL15 | 4772 | 1.121 | -0.1071 | No | ||
| 13 | CLTA | 4913 | 1.096 | -0.1068 | No | ||
| 14 | SGK | 4996 | 1.082 | -0.1034 | No | ||
| 15 | GNB2 | 5322 | 1.019 | -0.1136 | No | ||
| 16 | ARID5B | 5848 | 0.920 | -0.1354 | No | ||
| 17 | IRAK1 | 6121 | 0.869 | -0.1438 | No | ||
| 18 | PTPN23 | 6168 | 0.861 | -0.1401 | No | ||
| 19 | BACH2 | 6215 | 0.851 | -0.1364 | No | ||
| 20 | MLLT7 | 6713 | 0.761 | -0.1578 | No | ||
| 21 | RAB40C | 6753 | 0.754 | -0.1545 | No | ||
| 22 | BACH1 | 7275 | 0.667 | -0.1778 | No | ||
| 23 | TRPS1 | 7667 | 0.593 | -0.1946 | No | ||
| 24 | SNF1LK | 7841 | 0.559 | -0.2000 | No | ||
| 25 | ACVR2A | 7995 | 0.531 | -0.2044 | No | ||
| 26 | RNF31 | 8019 | 0.527 | -0.2018 | No | ||
| 27 | ITGAV | 8023 | 0.526 | -0.1982 | No | ||
| 28 | BTBD7 | 8375 | 0.461 | -0.2138 | No | ||
| 29 | UTY | 8385 | 0.460 | -0.2110 | No | ||
| 30 | EHF | 8823 | 0.378 | -0.2319 | No | ||
| 31 | TIPARP | 9234 | 0.291 | -0.2519 | No | ||
| 32 | CRTAM | 10531 | 0.014 | -0.3218 | No | ||
| 33 | SPRED1 | 10692 | -0.025 | -0.3303 | No | ||
| 34 | XPO1 | 10884 | -0.068 | -0.3401 | No | ||
| 35 | ATG16L1 | 11341 | -0.170 | -0.3635 | No | ||
| 36 | SOX11 | 11402 | -0.185 | -0.3654 | No | ||
| 37 | EGR2 | 11526 | -0.214 | -0.3705 | No | ||
| 38 | AFF2 | 11566 | -0.222 | -0.3710 | No | ||
| 39 | MGAT4A | 11922 | -0.304 | -0.3880 | No | ||
| 40 | GFI1 | 12138 | -0.356 | -0.3970 | No | ||
| 41 | PDE4B | 12666 | -0.489 | -0.4219 | No | ||
| 42 | TARDBP | 12728 | -0.503 | -0.4216 | No | ||
| 43 | MYST2 | 12861 | -0.536 | -0.4249 | No | ||
| 44 | INPP5A | 13133 | -0.612 | -0.4351 | No | ||
| 45 | ADCY9 | 13322 | -0.667 | -0.4405 | No | ||
| 46 | COPG | 13344 | -0.676 | -0.4367 | No | ||
| 47 | LRRC1 | 13413 | -0.694 | -0.4354 | No | ||
| 48 | VAMP3 | 13723 | -0.790 | -0.4464 | No | ||
| 49 | UTX | 13836 | -0.819 | -0.4465 | No | ||
| 50 | SLC7A11 | 14220 | -0.939 | -0.4605 | No | ||
| 51 | PURB | 14473 | -1.019 | -0.4667 | No | ||
| 52 | RGL2 | 14626 | -1.069 | -0.4672 | Yes | ||
| 53 | GTF2A1 | 14698 | -1.099 | -0.4632 | Yes | ||
| 54 | RAC1 | 14799 | -1.134 | -0.4604 | Yes | ||
| 55 | SYPL1 | 14808 | -1.138 | -0.4526 | Yes | ||
| 56 | HMGA2 | 15036 | -1.219 | -0.4561 | Yes | ||
| 57 | SNF1LK2 | 15291 | -1.323 | -0.4603 | Yes | ||
| 58 | EML4 | 15341 | -1.339 | -0.4533 | Yes | ||
| 59 | STX12 | 15354 | -1.343 | -0.4443 | Yes | ||
| 60 | ARNTL | 15540 | -1.419 | -0.4441 | Yes | ||
| 61 | PUM1 | 15776 | -1.521 | -0.4458 | Yes | ||
| 62 | SLCO4C1 | 15828 | -1.546 | -0.4374 | Yes | ||
| 63 | MAP3K7IP2 | 16007 | -1.630 | -0.4353 | Yes | ||
| 64 | RARG | 16032 | -1.643 | -0.4248 | Yes | ||
| 65 | FMNL2 | 16051 | -1.650 | -0.4139 | Yes | ||
| 66 | RERE | 16099 | -1.672 | -0.4044 | Yes | ||
| 67 | CCNT2 | 16150 | -1.693 | -0.3949 | Yes | ||
| 68 | MMD | 16151 | -1.693 | -0.3827 | Yes | ||
| 69 | ASB7 | 16249 | -1.732 | -0.3755 | Yes | ||
| 70 | FNDC3A | 16264 | -1.741 | -0.3637 | Yes | ||
| 71 | MORF4L1 | 16361 | -1.791 | -0.3560 | Yes | ||
| 72 | CFL2 | 16783 | -2.009 | -0.3643 | Yes | ||
| 73 | TGFBR1 | 16981 | -2.109 | -0.3597 | Yes | ||
| 74 | TIRAP | 17202 | -2.233 | -0.3555 | Yes | ||
| 75 | PSIP1 | 17253 | -2.263 | -0.3419 | Yes | ||
| 76 | BCLAF1 | 17319 | -2.299 | -0.3289 | Yes | ||
| 77 | MORF4L2 | 17449 | -2.380 | -0.3187 | Yes | ||
| 78 | ATF7IP | 17483 | -2.399 | -0.3032 | Yes | ||
| 79 | HMGB1 | 17643 | -2.502 | -0.2938 | Yes | ||
| 80 | STRN3 | 17765 | -2.578 | -0.2818 | Yes | ||
| 81 | ACSL4 | 17802 | -2.604 | -0.2650 | Yes | ||
| 82 | PREI3 | 17805 | -2.607 | -0.2463 | Yes | ||
| 83 | ERG | 18027 | -2.789 | -0.2382 | Yes | ||
| 84 | MARCKS | 18036 | -2.799 | -0.2185 | Yes | ||
| 85 | CSTF3 | 18086 | -2.844 | -0.2006 | Yes | ||
| 86 | STAU1 | 18093 | -2.851 | -0.1804 | Yes | ||
| 87 | DIRC2 | 18209 | -2.982 | -0.1652 | Yes | ||
| 88 | APC | 18346 | -3.202 | -0.1495 | Yes | ||
| 89 | ROCK2 | 18399 | -3.300 | -0.1285 | Yes | ||
| 90 | ATP2A2 | 18465 | -3.491 | -0.1069 | Yes | ||
| 91 | FBXO3 | 18516 | -3.701 | -0.0830 | Yes | ||
| 92 | PCAF | 18550 | -3.924 | -0.0565 | Yes | ||
| 93 | STAM | 18565 | -4.049 | -0.0281 | Yes | ||
| 94 | PPFIA1 | 18585 | -4.282 | 0.0017 | Yes |
