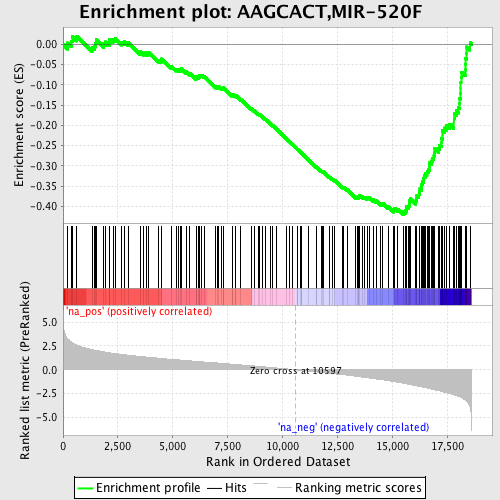
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | AAGCACT,MIR-520F |
| Enrichment Score (ES) | -0.4201272 |
| Normalized Enrichment Score (NES) | -2.0708277 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.621924E-4 |
| FWER p-Value | 0.01 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RKHD2 | 216 | 3.218 | 0.0033 | No | ||
| 2 | TOX | 386 | 2.899 | 0.0078 | No | ||
| 3 | RB1CC1 | 421 | 2.822 | 0.0191 | No | ||
| 4 | MNT | 624 | 2.589 | 0.0203 | No | ||
| 5 | AK2 | 1318 | 2.103 | -0.0074 | No | ||
| 6 | VANGL2 | 1440 | 2.045 | -0.0044 | No | ||
| 7 | VLDLR | 1482 | 2.031 | 0.0029 | No | ||
| 8 | KLHL18 | 1501 | 2.026 | 0.0114 | No | ||
| 9 | OLFM3 | 1851 | 1.884 | 0.0013 | No | ||
| 10 | HRBL | 1926 | 1.853 | 0.0059 | No | ||
| 11 | SPTLC1 | 2099 | 1.787 | 0.0050 | No | ||
| 12 | VSX1 | 2125 | 1.777 | 0.0119 | No | ||
| 13 | CREB5 | 2274 | 1.724 | 0.0120 | No | ||
| 14 | PAK7 | 2378 | 1.693 | 0.0143 | No | ||
| 15 | ATXN7L2 | 2677 | 1.613 | 0.0057 | No | ||
| 16 | CDC25B | 2787 | 1.580 | 0.0072 | No | ||
| 17 | MTMR3 | 2984 | 1.526 | 0.0037 | No | ||
| 18 | LETMD1 | 3513 | 1.395 | -0.0184 | No | ||
| 19 | MMP24 | 3668 | 1.359 | -0.0204 | No | ||
| 20 | EPAS1 | 3791 | 1.330 | -0.0208 | No | ||
| 21 | UHMK1 | 3897 | 1.305 | -0.0203 | No | ||
| 22 | ZBTB11 | 4369 | 1.207 | -0.0402 | No | ||
| 23 | CDKN1B | 4465 | 1.185 | -0.0398 | No | ||
| 24 | MTUS1 | 4499 | 1.177 | -0.0361 | No | ||
| 25 | ZNF385 | 4930 | 1.092 | -0.0543 | No | ||
| 26 | PRDM8 | 5146 | 1.053 | -0.0610 | No | ||
| 27 | CYP26B1 | 5258 | 1.032 | -0.0622 | No | ||
| 28 | FIGN | 5363 | 1.009 | -0.0631 | No | ||
| 29 | A2BP1 | 5385 | 1.005 | -0.0595 | No | ||
| 30 | OSBPL8 | 5626 | 0.960 | -0.0680 | No | ||
| 31 | ZFPM2 | 5771 | 0.934 | -0.0715 | No | ||
| 32 | CDH4 | 6058 | 0.882 | -0.0828 | No | ||
| 33 | RARB | 6070 | 0.879 | -0.0793 | No | ||
| 34 | LHX6 | 6157 | 0.863 | -0.0800 | No | ||
| 35 | BACH2 | 6215 | 0.851 | -0.0791 | No | ||
| 36 | WDR26 | 6230 | 0.849 | -0.0759 | No | ||
| 37 | ABCA1 | 6317 | 0.834 | -0.0766 | No | ||
| 38 | MUM1L1 | 6428 | 0.814 | -0.0788 | No | ||
| 39 | DLL4 | 6967 | 0.719 | -0.1045 | No | ||
| 40 | PLXNA1 | 7051 | 0.706 | -0.1057 | No | ||
| 41 | SLC24A3 | 7098 | 0.699 | -0.1050 | No | ||
| 42 | WNT5A | 7213 | 0.677 | -0.1080 | No | ||
| 43 | CXXC5 | 7290 | 0.666 | -0.1090 | No | ||
| 44 | ASF1B | 7304 | 0.664 | -0.1066 | No | ||
| 45 | RAP1A | 7716 | 0.583 | -0.1261 | No | ||
| 46 | NTN4 | 7735 | 0.579 | -0.1244 | No | ||
| 47 | SNF1LK | 7841 | 0.559 | -0.1275 | No | ||
| 48 | MKRN1 | 7861 | 0.556 | -0.1259 | No | ||
| 49 | RFX4 | 8101 | 0.511 | -0.1364 | No | ||
| 50 | MEF2C | 8568 | 0.422 | -0.1597 | No | ||
| 51 | NFIB | 8575 | 0.420 | -0.1581 | No | ||
| 52 | HN1 | 8714 | 0.397 | -0.1637 | No | ||
| 53 | LEFTY1 | 8906 | 0.362 | -0.1723 | No | ||
| 54 | XYLT1 | 8972 | 0.350 | -0.1742 | No | ||
| 55 | APP | 9071 | 0.328 | -0.1780 | No | ||
| 56 | TIPARP | 9234 | 0.291 | -0.1854 | No | ||
| 57 | RGMA | 9443 | 0.245 | -0.1955 | No | ||
| 58 | ESR1 | 9562 | 0.220 | -0.2009 | No | ||
| 59 | PANK2 | 9724 | 0.184 | -0.2088 | No | ||
| 60 | ZKSCAN1 | 10169 | 0.092 | -0.2324 | No | ||
| 61 | NR2C2 | 10323 | 0.059 | -0.2404 | No | ||
| 62 | PAPOLA | 10461 | 0.030 | -0.2477 | No | ||
| 63 | CCND2 | 10685 | -0.024 | -0.2596 | No | ||
| 64 | SPRED1 | 10692 | -0.025 | -0.2598 | No | ||
| 65 | H2AFY | 10820 | -0.055 | -0.2665 | No | ||
| 66 | TNFAIP1 | 10853 | -0.061 | -0.2679 | No | ||
| 67 | FBXW11 | 10879 | -0.066 | -0.2689 | No | ||
| 68 | SF4 | 11174 | -0.134 | -0.2842 | No | ||
| 69 | KCNMA1 | 11545 | -0.217 | -0.3033 | No | ||
| 70 | ARID4B | 11551 | -0.218 | -0.3025 | No | ||
| 71 | PGBD5 | 11771 | -0.269 | -0.3131 | No | ||
| 72 | HSPA14 | 11826 | -0.281 | -0.3147 | No | ||
| 73 | COL4A3 | 11850 | -0.288 | -0.3146 | No | ||
| 74 | PHF15 | 12120 | -0.351 | -0.3276 | No | ||
| 75 | TUBG1 | 12162 | -0.363 | -0.3281 | No | ||
| 76 | CREBL1 | 12279 | -0.389 | -0.3326 | No | ||
| 77 | FGD5 | 12381 | -0.414 | -0.3361 | No | ||
| 78 | TARDBP | 12728 | -0.503 | -0.3525 | No | ||
| 79 | EMX2 | 12783 | -0.515 | -0.3530 | No | ||
| 80 | UBE2B | 12943 | -0.557 | -0.3590 | No | ||
| 81 | PLAG1 | 13330 | -0.670 | -0.3768 | No | ||
| 82 | RGL1 | 13418 | -0.695 | -0.3782 | No | ||
| 83 | ADCYAP1 | 13480 | -0.712 | -0.3782 | No | ||
| 84 | SLC6A9 | 13481 | -0.712 | -0.3749 | No | ||
| 85 | NEK9 | 13498 | -0.717 | -0.3724 | No | ||
| 86 | DPP4 | 13644 | -0.765 | -0.3767 | No | ||
| 87 | VAMP3 | 13723 | -0.790 | -0.3772 | No | ||
| 88 | NR4A3 | 13850 | -0.823 | -0.3802 | No | ||
| 89 | IL6ST | 13872 | -0.828 | -0.3774 | No | ||
| 90 | SEPT2 | 13960 | -0.851 | -0.3782 | No | ||
| 91 | ZNF462 | 14133 | -0.913 | -0.3832 | No | ||
| 92 | ARID4A | 14265 | -0.953 | -0.3859 | No | ||
| 93 | PURB | 14473 | -1.019 | -0.3923 | No | ||
| 94 | RASSF2 | 14558 | -1.042 | -0.3920 | No | ||
| 95 | ITGA9 | 14826 | -1.143 | -0.4011 | No | ||
| 96 | IQWD1 | 15067 | -1.232 | -0.4083 | No | ||
| 97 | ARHGEF17 | 15112 | -1.254 | -0.4049 | No | ||
| 98 | MAP3K14 | 15259 | -1.311 | -0.4066 | No | ||
| 99 | PBX3 | 15509 | -1.408 | -0.4135 | Yes | ||
| 100 | HIVEP3 | 15590 | -1.443 | -0.4111 | Yes | ||
| 101 | EDNRB | 15644 | -1.464 | -0.4072 | Yes | ||
| 102 | TMEM16F | 15659 | -1.473 | -0.4010 | Yes | ||
| 103 | DDHD1 | 15761 | -1.516 | -0.3994 | Yes | ||
| 104 | PICALM | 15769 | -1.519 | -0.3927 | Yes | ||
| 105 | AEBP2 | 15783 | -1.525 | -0.3863 | Yes | ||
| 106 | CPD | 15815 | -1.539 | -0.3807 | Yes | ||
| 107 | MINK1 | 16070 | -1.658 | -0.3868 | Yes | ||
| 108 | ATP2B2 | 16115 | -1.679 | -0.3813 | Yes | ||
| 109 | DDX3X | 16124 | -1.681 | -0.3739 | Yes | ||
| 110 | SLC38A2 | 16220 | -1.723 | -0.3709 | Yes | ||
| 111 | SS18L1 | 16235 | -1.729 | -0.3636 | Yes | ||
| 112 | FNDC3A | 16264 | -1.741 | -0.3570 | Yes | ||
| 113 | SAR1B | 16325 | -1.775 | -0.3519 | Yes | ||
| 114 | RABEP1 | 16350 | -1.788 | -0.3449 | Yes | ||
| 115 | ASF1A | 16393 | -1.808 | -0.3387 | Yes | ||
| 116 | ABR | 16424 | -1.822 | -0.3318 | Yes | ||
| 117 | IRF2 | 16448 | -1.830 | -0.3245 | Yes | ||
| 118 | ATAD2 | 16512 | -1.861 | -0.3192 | Yes | ||
| 119 | CXXC6 | 16586 | -1.901 | -0.3143 | Yes | ||
| 120 | SENP1 | 16642 | -1.931 | -0.3082 | Yes | ||
| 121 | DNAJC13 | 16694 | -1.955 | -0.3018 | Yes | ||
| 122 | GAB2 | 16699 | -1.959 | -0.2929 | Yes | ||
| 123 | CFL2 | 16783 | -2.009 | -0.2880 | Yes | ||
| 124 | RDX | 16824 | -2.031 | -0.2807 | Yes | ||
| 125 | NT5C3 | 16903 | -2.067 | -0.2752 | Yes | ||
| 126 | RNF6 | 16916 | -2.073 | -0.2662 | Yes | ||
| 127 | RPS6KA5 | 16943 | -2.089 | -0.2578 | Yes | ||
| 128 | YTHDF3 | 17117 | -2.184 | -0.2570 | Yes | ||
| 129 | MBNL2 | 17164 | -2.211 | -0.2491 | Yes | ||
| 130 | AP3M1 | 17244 | -2.256 | -0.2429 | Yes | ||
| 131 | PTPLB | 17251 | -2.261 | -0.2326 | Yes | ||
| 132 | MTCH2 | 17292 | -2.286 | -0.2241 | Yes | ||
| 133 | FLT1 | 17293 | -2.287 | -0.2134 | Yes | ||
| 134 | ARF1 | 17369 | -2.331 | -0.2066 | Yes | ||
| 135 | RAB22A | 17466 | -2.387 | -0.2006 | Yes | ||
| 136 | PRC1 | 17622 | -2.488 | -0.1974 | Yes | ||
| 137 | ZFYVE26 | 17814 | -2.612 | -0.1955 | Yes | ||
| 138 | RBL1 | 17815 | -2.613 | -0.1833 | Yes | ||
| 139 | CDH2 | 17829 | -2.624 | -0.1717 | Yes | ||
| 140 | CUGBP2 | 17921 | -2.698 | -0.1640 | Yes | ||
| 141 | PRKACB | 18025 | -2.788 | -0.1566 | Yes | ||
| 142 | FRAS1 | 18072 | -2.831 | -0.1458 | Yes | ||
| 143 | RABGAP1 | 18084 | -2.842 | -0.1331 | Yes | ||
| 144 | KPNB1 | 18123 | -2.884 | -0.1217 | Yes | ||
| 145 | SYNC1 | 18128 | -2.897 | -0.1084 | Yes | ||
| 146 | RALGDS | 18129 | -2.897 | -0.0948 | Yes | ||
| 147 | SLITRK3 | 18135 | -2.904 | -0.0815 | Yes | ||
| 148 | SP1 | 18146 | -2.912 | -0.0684 | Yes | ||
| 149 | RAB11A | 18323 | -3.164 | -0.0632 | Yes | ||
| 150 | CDCA7 | 18329 | -3.166 | -0.0487 | Yes | ||
| 151 | DNAJA2 | 18353 | -3.221 | -0.0348 | Yes | ||
| 152 | MECP2 | 18396 | -3.287 | -0.0217 | Yes | ||
| 153 | WDR37 | 18402 | -3.309 | -0.0065 | Yes | ||
| 154 | CCND1 | 18545 | -3.865 | 0.0038 | Yes |
