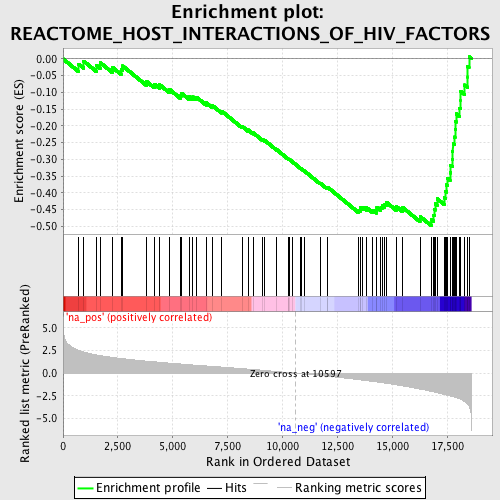
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS |
| Enrichment Score (ES) | -0.49900374 |
| Normalized Enrichment Score (NES) | -2.1659603 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0035662984 |
| FWER p-Value | 0.019 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AP2B1 | 708 | 2.504 | -0.0154 | No | ||
| 2 | BANF1 | 939 | 2.326 | -0.0066 | No | ||
| 3 | FYN | 1528 | 2.012 | -0.0200 | No | ||
| 4 | PSMD9 | 1691 | 1.948 | -0.0110 | No | ||
| 5 | NUP188 | 2241 | 1.736 | -0.0248 | No | ||
| 6 | NUP62 | 2647 | 1.622 | -0.0319 | No | ||
| 7 | PSMB2 | 2714 | 1.601 | -0.0209 | No | ||
| 8 | PACS1 | 3804 | 1.328 | -0.0675 | No | ||
| 9 | PSMB10 | 4161 | 1.254 | -0.0753 | No | ||
| 10 | AP2A2 | 4398 | 1.200 | -0.0771 | No | ||
| 11 | TCEB2 | 4852 | 1.106 | -0.0914 | No | ||
| 12 | PSMC4 | 5367 | 1.009 | -0.1100 | No | ||
| 13 | LCK | 5387 | 1.005 | -0.1018 | No | ||
| 14 | PSMB6 | 5748 | 0.939 | -0.1127 | No | ||
| 15 | PSMA7 | 5916 | 0.906 | -0.1135 | No | ||
| 16 | PSMD5 | 6081 | 0.877 | -0.1143 | No | ||
| 17 | B2M | 6524 | 0.797 | -0.1309 | No | ||
| 18 | PSMC1 | 6801 | 0.743 | -0.1390 | No | ||
| 19 | CD28 | 7225 | 0.675 | -0.1557 | No | ||
| 20 | PSMA5 | 8161 | 0.498 | -0.2016 | No | ||
| 21 | HCK | 8431 | 0.449 | -0.2120 | No | ||
| 22 | PSMB3 | 8664 | 0.405 | -0.2208 | No | ||
| 23 | NUP35 | 9086 | 0.325 | -0.2406 | No | ||
| 24 | AP1B1 | 9199 | 0.298 | -0.2439 | No | ||
| 25 | PSMB4 | 9741 | 0.181 | -0.2714 | No | ||
| 26 | PSMB9 | 10277 | 0.070 | -0.2997 | No | ||
| 27 | PSMB5 | 10292 | 0.067 | -0.2998 | No | ||
| 28 | PSMA6 | 10299 | 0.065 | -0.2995 | No | ||
| 29 | CDK9 | 10456 | 0.030 | -0.3077 | No | ||
| 30 | PSMD8 | 10838 | -0.059 | -0.3277 | No | ||
| 31 | XPO1 | 10884 | -0.068 | -0.3295 | No | ||
| 32 | PSMC3 | 10997 | -0.093 | -0.3347 | No | ||
| 33 | PSMC2 | 11717 | -0.255 | -0.3711 | No | ||
| 34 | PSMB1 | 12039 | -0.334 | -0.3854 | No | ||
| 35 | RAE1 | 12061 | -0.339 | -0.3835 | No | ||
| 36 | RAN | 13479 | -0.712 | -0.4534 | No | ||
| 37 | PSMD2 | 13539 | -0.730 | -0.4499 | No | ||
| 38 | PSME3 | 13560 | -0.736 | -0.4443 | No | ||
| 39 | PPIA | 13664 | -0.770 | -0.4429 | No | ||
| 40 | NUPL2 | 13833 | -0.818 | -0.4445 | No | ||
| 41 | NUP43 | 14122 | -0.908 | -0.4517 | No | ||
| 42 | PSME1 | 14286 | -0.960 | -0.4518 | No | ||
| 43 | NUP153 | 14299 | -0.964 | -0.4437 | No | ||
| 44 | PSMC5 | 14485 | -1.021 | -0.4443 | No | ||
| 45 | PSMD11 | 14541 | -1.037 | -0.4379 | No | ||
| 46 | PSMA1 | 14663 | -1.082 | -0.4345 | No | ||
| 47 | RANGAP1 | 14734 | -1.109 | -0.4282 | No | ||
| 48 | PSMC6 | 15180 | -1.279 | -0.4406 | No | ||
| 49 | RANBP1 | 15480 | -1.397 | -0.4440 | No | ||
| 50 | NUP155 | 16271 | -1.744 | -0.4707 | No | ||
| 51 | NUP37 | 16797 | -2.018 | -0.4806 | Yes | ||
| 52 | PSMD3 | 16879 | -2.057 | -0.4663 | Yes | ||
| 53 | PAK2 | 16919 | -2.075 | -0.4495 | Yes | ||
| 54 | NUP160 | 16956 | -2.095 | -0.4323 | Yes | ||
| 55 | RANBP2 | 17048 | -2.144 | -0.4177 | Yes | ||
| 56 | ARF1 | 17369 | -2.331 | -0.4138 | Yes | ||
| 57 | NUP93 | 17425 | -2.364 | -0.3952 | Yes | ||
| 58 | PSMD4 | 17477 | -2.395 | -0.3761 | Yes | ||
| 59 | PSMD7 | 17529 | -2.431 | -0.3568 | Yes | ||
| 60 | CCNT1 | 17639 | -2.498 | -0.3399 | Yes | ||
| 61 | PSMA4 | 17660 | -2.511 | -0.3181 | Yes | ||
| 62 | PSMD10 | 17742 | -2.569 | -0.2991 | Yes | ||
| 63 | NUP54 | 17751 | -2.571 | -0.2761 | Yes | ||
| 64 | PSMA2 | 17770 | -2.581 | -0.2536 | Yes | ||
| 65 | RBX1 | 17836 | -2.628 | -0.2331 | Yes | ||
| 66 | PSMA3 | 17865 | -2.659 | -0.2104 | Yes | ||
| 67 | KPNA1 | 17888 | -2.676 | -0.1873 | Yes | ||
| 68 | TPR | 17914 | -2.694 | -0.1641 | Yes | ||
| 69 | TCEB1 | 18070 | -2.831 | -0.1467 | Yes | ||
| 70 | NUP107 | 18117 | -2.875 | -0.1230 | Yes | ||
| 71 | KPNB1 | 18123 | -2.884 | -0.0970 | Yes | ||
| 72 | PSMD14 | 18296 | -3.126 | -0.0778 | Yes | ||
| 73 | PSMD12 | 18428 | -3.388 | -0.0540 | Yes | ||
| 74 | CD4 | 18432 | -3.407 | -0.0231 | Yes | ||
| 75 | AP1G1 | 18502 | -3.623 | 0.0061 | Yes |
