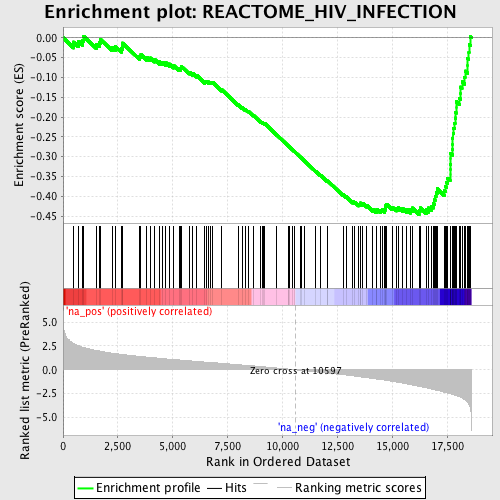
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_HIV_INFECTION |
| Enrichment Score (ES) | -0.4461138 |
| Normalized Enrichment Score (NES) | -2.1604626 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0030720332 |
| FWER p-Value | 0.021 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RDBP | 477 | 2.753 | -0.0106 | No | ||
| 2 | AP2B1 | 708 | 2.504 | -0.0092 | No | ||
| 3 | ERCC2 | 889 | 2.361 | -0.0060 | No | ||
| 4 | BANF1 | 939 | 2.326 | 0.0042 | No | ||
| 5 | FYN | 1528 | 2.012 | -0.0165 | No | ||
| 6 | TAF10 | 1650 | 1.962 | -0.0122 | No | ||
| 7 | PSMD9 | 1691 | 1.948 | -0.0036 | No | ||
| 8 | NUP188 | 2241 | 1.736 | -0.0237 | No | ||
| 9 | POLR2E | 2385 | 1.691 | -0.0221 | No | ||
| 10 | NUP62 | 2647 | 1.622 | -0.0273 | No | ||
| 11 | POLR2B | 2703 | 1.604 | -0.0214 | No | ||
| 12 | PSMB2 | 2714 | 1.601 | -0.0131 | No | ||
| 13 | LIG1 | 3491 | 1.400 | -0.0474 | No | ||
| 14 | TAF6 | 3546 | 1.388 | -0.0426 | No | ||
| 15 | PACS1 | 3804 | 1.328 | -0.0492 | No | ||
| 16 | TH1L | 3968 | 1.294 | -0.0509 | No | ||
| 17 | PSMB10 | 4161 | 1.254 | -0.0544 | No | ||
| 18 | AP2A2 | 4398 | 1.200 | -0.0605 | No | ||
| 19 | SUPT4H1 | 4536 | 1.168 | -0.0615 | No | ||
| 20 | TAF12 | 4687 | 1.139 | -0.0633 | No | ||
| 21 | TCEB2 | 4852 | 1.106 | -0.0661 | No | ||
| 22 | POLR2H | 5044 | 1.073 | -0.0705 | No | ||
| 23 | POLR2G | 5288 | 1.026 | -0.0780 | No | ||
| 24 | PSMC4 | 5367 | 1.009 | -0.0766 | No | ||
| 25 | LCK | 5387 | 1.005 | -0.0721 | No | ||
| 26 | PSMB6 | 5748 | 0.939 | -0.0864 | No | ||
| 27 | PSMA7 | 5916 | 0.906 | -0.0904 | No | ||
| 28 | PSMD5 | 6081 | 0.877 | -0.0945 | No | ||
| 29 | CDK7 | 6460 | 0.809 | -0.1104 | No | ||
| 30 | B2M | 6524 | 0.797 | -0.1095 | No | ||
| 31 | POLR2I | 6639 | 0.776 | -0.1113 | No | ||
| 32 | GTF2A2 | 6730 | 0.758 | -0.1120 | No | ||
| 33 | PSMC1 | 6801 | 0.743 | -0.1117 | No | ||
| 34 | CD28 | 7225 | 0.675 | -0.1309 | No | ||
| 35 | POLR2J | 7990 | 0.533 | -0.1692 | No | ||
| 36 | PSMA5 | 8161 | 0.498 | -0.1757 | No | ||
| 37 | XRCC5 | 8298 | 0.473 | -0.1804 | No | ||
| 38 | HCK | 8431 | 0.449 | -0.1851 | No | ||
| 39 | PSMB3 | 8664 | 0.405 | -0.1954 | No | ||
| 40 | ELL | 9004 | 0.343 | -0.2118 | No | ||
| 41 | NUP35 | 9086 | 0.325 | -0.2144 | No | ||
| 42 | GTF2H3 | 9134 | 0.315 | -0.2152 | No | ||
| 43 | AP1B1 | 9199 | 0.298 | -0.2170 | No | ||
| 44 | PSMB4 | 9741 | 0.181 | -0.2453 | No | ||
| 45 | PSMB9 | 10277 | 0.070 | -0.2738 | No | ||
| 46 | PSMB5 | 10292 | 0.067 | -0.2742 | No | ||
| 47 | PSMA6 | 10299 | 0.065 | -0.2742 | No | ||
| 48 | CDK9 | 10456 | 0.030 | -0.2824 | No | ||
| 49 | POLR2A | 10546 | 0.010 | -0.2872 | No | ||
| 50 | PSMD8 | 10838 | -0.059 | -0.3026 | No | ||
| 51 | XPO1 | 10884 | -0.068 | -0.3047 | No | ||
| 52 | PSMC3 | 10997 | -0.093 | -0.3102 | No | ||
| 53 | TAF4 | 11511 | -0.211 | -0.3368 | No | ||
| 54 | PSMC2 | 11717 | -0.255 | -0.3465 | No | ||
| 55 | PSMB1 | 12039 | -0.334 | -0.3620 | No | ||
| 56 | RAE1 | 12061 | -0.339 | -0.3613 | No | ||
| 57 | GTF2F1 | 12759 | -0.510 | -0.3961 | No | ||
| 58 | TCEB3 | 12896 | -0.544 | -0.4005 | No | ||
| 59 | FEN1 | 13207 | -0.630 | -0.4138 | No | ||
| 60 | TAF9 | 13299 | -0.660 | -0.4150 | No | ||
| 61 | RAN | 13479 | -0.712 | -0.4208 | No | ||
| 62 | PSMD2 | 13539 | -0.730 | -0.4200 | No | ||
| 63 | PSME3 | 13560 | -0.736 | -0.4170 | No | ||
| 64 | PPIA | 13664 | -0.770 | -0.4183 | No | ||
| 65 | NUPL2 | 13833 | -0.818 | -0.4229 | No | ||
| 66 | NUP43 | 14122 | -0.908 | -0.4334 | No | ||
| 67 | PSME1 | 14286 | -0.960 | -0.4370 | Yes | ||
| 68 | NUP153 | 14299 | -0.964 | -0.4323 | Yes | ||
| 69 | PSMC5 | 14485 | -1.021 | -0.4367 | Yes | ||
| 70 | PSMD11 | 14541 | -1.037 | -0.4339 | Yes | ||
| 71 | PSMA1 | 14663 | -1.082 | -0.4345 | Yes | ||
| 72 | ERCC3 | 14676 | -1.088 | -0.4291 | Yes | ||
| 73 | GTF2A1 | 14698 | -1.099 | -0.4242 | Yes | ||
| 74 | RANGAP1 | 14734 | -1.109 | -0.4200 | Yes | ||
| 75 | GTF2F2 | 15008 | -1.211 | -0.4281 | Yes | ||
| 76 | PSMC6 | 15180 | -1.279 | -0.4303 | Yes | ||
| 77 | MNAT1 | 15294 | -1.324 | -0.4291 | Yes | ||
| 78 | RANBP1 | 15480 | -1.397 | -0.4314 | Yes | ||
| 79 | POLR2C | 15650 | -1.466 | -0.4324 | Yes | ||
| 80 | NCBP2 | 15840 | -1.552 | -0.4341 | Yes | ||
| 81 | TAF11 | 15919 | -1.587 | -0.4296 | Yes | ||
| 82 | XRCC4 | 16226 | -1.726 | -0.4366 | Yes | ||
| 83 | NUP155 | 16271 | -1.744 | -0.4294 | Yes | ||
| 84 | TBP | 16552 | -1.880 | -0.4341 | Yes | ||
| 85 | LIG4 | 16666 | -1.938 | -0.4296 | Yes | ||
| 86 | NUP37 | 16797 | -2.018 | -0.4255 | Yes | ||
| 87 | PSMD3 | 16879 | -2.057 | -0.4185 | Yes | ||
| 88 | PAK2 | 16919 | -2.075 | -0.4092 | Yes | ||
| 89 | NUP160 | 16956 | -2.095 | -0.3996 | Yes | ||
| 90 | RNMT | 17000 | -2.120 | -0.3902 | Yes | ||
| 91 | RANBP2 | 17048 | -2.144 | -0.3809 | Yes | ||
| 92 | ARF1 | 17369 | -2.331 | -0.3854 | Yes | ||
| 93 | NUP93 | 17425 | -2.364 | -0.3753 | Yes | ||
| 94 | PSMD4 | 17477 | -2.395 | -0.3649 | Yes | ||
| 95 | PSMD7 | 17529 | -2.431 | -0.3542 | Yes | ||
| 96 | CCNT1 | 17639 | -2.498 | -0.3463 | Yes | ||
| 97 | XRCC6 | 17640 | -2.499 | -0.3325 | Yes | ||
| 98 | GTF2H1 | 17644 | -2.502 | -0.3189 | Yes | ||
| 99 | GTF2E2 | 17658 | -2.510 | -0.3058 | Yes | ||
| 100 | PSMA4 | 17660 | -2.511 | -0.2920 | Yes | ||
| 101 | PSMD10 | 17742 | -2.569 | -0.2822 | Yes | ||
| 102 | NUP54 | 17751 | -2.571 | -0.2685 | Yes | ||
| 103 | RNGTT | 17757 | -2.575 | -0.2545 | Yes | ||
| 104 | PSMA2 | 17770 | -2.581 | -0.2409 | Yes | ||
| 105 | GTF2H2 | 17799 | -2.602 | -0.2281 | Yes | ||
| 106 | RBX1 | 17836 | -2.628 | -0.2156 | Yes | ||
| 107 | PSMA3 | 17865 | -2.659 | -0.2024 | Yes | ||
| 108 | KPNA1 | 17888 | -2.676 | -0.1888 | Yes | ||
| 109 | GTF2E1 | 17912 | -2.692 | -0.1752 | Yes | ||
| 110 | TPR | 17914 | -2.694 | -0.1604 | Yes | ||
| 111 | TCEB1 | 18070 | -2.831 | -0.1532 | Yes | ||
| 112 | NUP107 | 18117 | -2.875 | -0.1398 | Yes | ||
| 113 | KPNB1 | 18123 | -2.884 | -0.1242 | Yes | ||
| 114 | TAF13 | 18182 | -2.948 | -0.1111 | Yes | ||
| 115 | PSMD14 | 18296 | -3.126 | -0.1000 | Yes | ||
| 116 | CCNH | 18320 | -3.158 | -0.0838 | Yes | ||
| 117 | PSMD12 | 18428 | -3.388 | -0.0709 | Yes | ||
| 118 | CD4 | 18432 | -3.407 | -0.0523 | Yes | ||
| 119 | GTF2B | 18495 | -3.569 | -0.0359 | Yes | ||
| 120 | AP1G1 | 18502 | -3.623 | -0.0163 | Yes | ||
| 121 | TAF5 | 18566 | -4.062 | 0.0027 | Yes |
