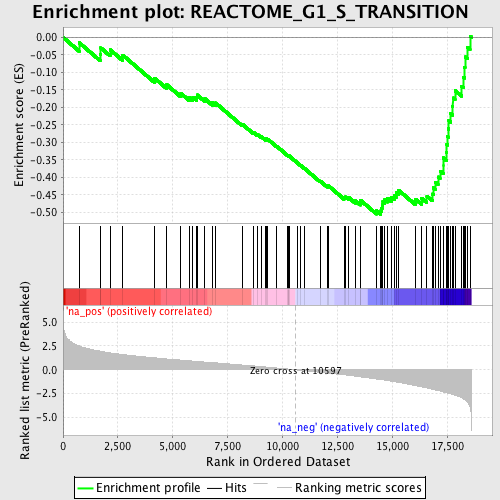
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_G1_S_TRANSITION |
| Enrichment Score (ES) | -0.5059156 |
| Normalized Enrichment Score (NES) | -2.2342417 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0039882422 |
| FWER p-Value | 0.0060 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TK2 | 732 | 2.480 | -0.0151 | No | ||
| 2 | PSMD9 | 1691 | 1.948 | -0.0476 | No | ||
| 3 | RPA2 | 1719 | 1.935 | -0.0300 | No | ||
| 4 | CCNE1 | 2145 | 1.770 | -0.0355 | No | ||
| 5 | PSMB2 | 2714 | 1.601 | -0.0504 | No | ||
| 6 | PSMB10 | 4161 | 1.254 | -0.1161 | No | ||
| 7 | MCM7 | 4727 | 1.130 | -0.1354 | No | ||
| 8 | PSMC4 | 5367 | 1.009 | -0.1600 | No | ||
| 9 | PSMB6 | 5748 | 0.939 | -0.1712 | No | ||
| 10 | PSMA7 | 5916 | 0.906 | -0.1713 | No | ||
| 11 | PSMD5 | 6081 | 0.877 | -0.1715 | No | ||
| 12 | TYMS | 6108 | 0.872 | -0.1644 | No | ||
| 13 | CDK7 | 6460 | 0.809 | -0.1753 | No | ||
| 14 | PSMC1 | 6801 | 0.743 | -0.1864 | No | ||
| 15 | POLE2 | 6949 | 0.721 | -0.1872 | No | ||
| 16 | PSMA5 | 8161 | 0.498 | -0.2476 | No | ||
| 17 | PSMB3 | 8664 | 0.405 | -0.2707 | No | ||
| 18 | POLA1 | 8844 | 0.374 | -0.2767 | No | ||
| 19 | MCM5 | 9024 | 0.339 | -0.2830 | No | ||
| 20 | MCM2 | 9236 | 0.291 | -0.2915 | No | ||
| 21 | E2F1 | 9255 | 0.288 | -0.2896 | No | ||
| 22 | POLA2 | 9324 | 0.275 | -0.2906 | No | ||
| 23 | PSMB4 | 9741 | 0.181 | -0.3113 | No | ||
| 24 | MCM3 | 10237 | 0.078 | -0.3372 | No | ||
| 25 | PSMB9 | 10277 | 0.070 | -0.3386 | No | ||
| 26 | PSMB5 | 10292 | 0.067 | -0.3387 | No | ||
| 27 | PSMA6 | 10299 | 0.065 | -0.3384 | No | ||
| 28 | CCNA1 | 10686 | -0.025 | -0.3590 | No | ||
| 29 | PSMD8 | 10838 | -0.059 | -0.3665 | No | ||
| 30 | PSMC3 | 10997 | -0.093 | -0.3741 | No | ||
| 31 | PSMC2 | 11717 | -0.255 | -0.4104 | No | ||
| 32 | PSMB1 | 12039 | -0.334 | -0.4244 | No | ||
| 33 | MCM10 | 12091 | -0.345 | -0.4238 | No | ||
| 34 | CDKN1A | 12803 | -0.522 | -0.4570 | No | ||
| 35 | DHFR | 12879 | -0.540 | -0.4557 | No | ||
| 36 | CDC25A | 13018 | -0.579 | -0.4575 | No | ||
| 37 | ORC2L | 13315 | -0.665 | -0.4669 | No | ||
| 38 | PSMD2 | 13539 | -0.730 | -0.4717 | No | ||
| 39 | PSME3 | 13560 | -0.736 | -0.4656 | No | ||
| 40 | PSME1 | 14286 | -0.960 | -0.4952 | No | ||
| 41 | PSMC5 | 14485 | -1.021 | -0.4959 | Yes | ||
| 42 | ORC1L | 14523 | -1.032 | -0.4877 | Yes | ||
| 43 | PSMD11 | 14541 | -1.037 | -0.4784 | Yes | ||
| 44 | TFDP1 | 14575 | -1.050 | -0.4699 | Yes | ||
| 45 | PSMA1 | 14663 | -1.082 | -0.4639 | Yes | ||
| 46 | WEE1 | 14784 | -1.131 | -0.4593 | Yes | ||
| 47 | E2F2 | 14975 | -1.199 | -0.4577 | Yes | ||
| 48 | MCM6 | 15098 | -1.246 | -0.4520 | Yes | ||
| 49 | PSMC6 | 15180 | -1.279 | -0.4438 | Yes | ||
| 50 | MNAT1 | 15294 | -1.324 | -0.4369 | Yes | ||
| 51 | RPA3 | 16062 | -1.655 | -0.4620 | Yes | ||
| 52 | ORC5L | 16341 | -1.784 | -0.4594 | Yes | ||
| 53 | CDC45L | 16582 | -1.897 | -0.4537 | Yes | ||
| 54 | CUL1 | 16836 | -2.035 | -0.4473 | Yes | ||
| 55 | PSMD3 | 16879 | -2.057 | -0.4294 | Yes | ||
| 56 | PRIM1 | 16962 | -2.096 | -0.4132 | Yes | ||
| 57 | PCNA | 17091 | -2.167 | -0.3988 | Yes | ||
| 58 | RPA1 | 17181 | -2.221 | -0.3817 | Yes | ||
| 59 | RB1 | 17323 | -2.301 | -0.3667 | Yes | ||
| 60 | RRM2 | 17325 | -2.302 | -0.3441 | Yes | ||
| 61 | PSMD4 | 17477 | -2.395 | -0.3287 | Yes | ||
| 62 | CDK2 | 17488 | -2.401 | -0.3056 | Yes | ||
| 63 | PSMD7 | 17529 | -2.431 | -0.2838 | Yes | ||
| 64 | CCNB1 | 17554 | -2.442 | -0.2611 | Yes | ||
| 65 | POLE | 17578 | -2.455 | -0.2382 | Yes | ||
| 66 | PSMA4 | 17660 | -2.511 | -0.2178 | Yes | ||
| 67 | PSMD10 | 17742 | -2.569 | -0.1969 | Yes | ||
| 68 | PSMA2 | 17770 | -2.581 | -0.1730 | Yes | ||
| 69 | PSMA3 | 17865 | -2.659 | -0.1519 | Yes | ||
| 70 | CDC7 | 18162 | -2.928 | -0.1391 | Yes | ||
| 71 | MCM4 | 18238 | -3.033 | -0.1133 | Yes | ||
| 72 | PSMD14 | 18296 | -3.126 | -0.0856 | Yes | ||
| 73 | CCNH | 18320 | -3.158 | -0.0558 | Yes | ||
| 74 | PSMD12 | 18428 | -3.388 | -0.0282 | Yes | ||
| 75 | ORC4L | 18548 | -3.893 | 0.0037 | Yes |
