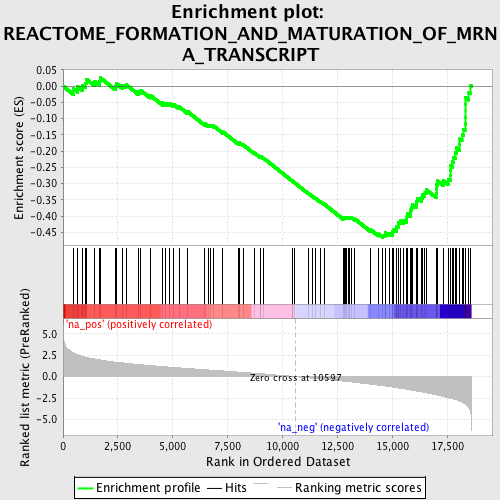
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | -0.46554667 |
| Normalized Enrichment Score (NES) | -2.1640692 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0032882742 |
| FWER p-Value | 0.02 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RDBP | 477 | 2.753 | -0.0080 | No | ||
| 2 | SF3A2 | 668 | 2.545 | -0.0019 | No | ||
| 3 | ERCC2 | 889 | 2.361 | 0.0015 | No | ||
| 4 | SNRPD2 | 1010 | 2.276 | 0.0097 | No | ||
| 5 | SNRPA | 1053 | 2.253 | 0.0219 | No | ||
| 6 | SNRPB | 1442 | 2.044 | 0.0141 | No | ||
| 7 | TAF10 | 1650 | 1.962 | 0.0156 | No | ||
| 8 | U2AF2 | 1684 | 1.950 | 0.0264 | No | ||
| 9 | POLR2E | 2385 | 1.691 | -0.0005 | No | ||
| 10 | CD2BP2 | 2424 | 1.680 | 0.0083 | No | ||
| 11 | POLR2B | 2703 | 1.604 | 0.0036 | No | ||
| 12 | SRRM1 | 2869 | 1.556 | 0.0047 | No | ||
| 13 | SNRPE | 3431 | 1.411 | -0.0165 | No | ||
| 14 | TAF6 | 3546 | 1.388 | -0.0137 | No | ||
| 15 | TH1L | 3968 | 1.294 | -0.0281 | No | ||
| 16 | SUPT4H1 | 4536 | 1.168 | -0.0512 | No | ||
| 17 | TAF12 | 4687 | 1.139 | -0.0520 | No | ||
| 18 | TCEB2 | 4852 | 1.106 | -0.0537 | No | ||
| 19 | POLR2H | 5044 | 1.073 | -0.0571 | No | ||
| 20 | POLR2G | 5288 | 1.026 | -0.0636 | No | ||
| 21 | RNPS1 | 5670 | 0.953 | -0.0780 | No | ||
| 22 | CDK7 | 6460 | 0.809 | -0.1154 | No | ||
| 23 | POLR2I | 6639 | 0.776 | -0.1200 | No | ||
| 24 | GTF2A2 | 6730 | 0.758 | -0.1200 | No | ||
| 25 | TXNL4A | 6847 | 0.735 | -0.1215 | No | ||
| 26 | NUDT21 | 7286 | 0.666 | -0.1409 | No | ||
| 27 | POLR2J | 7990 | 0.533 | -0.1754 | No | ||
| 28 | U2AF1 | 8030 | 0.525 | -0.1742 | No | ||
| 29 | SF3B4 | 8215 | 0.489 | -0.1809 | No | ||
| 30 | NFX1 | 8716 | 0.396 | -0.2054 | No | ||
| 31 | SF3B1 | 8984 | 0.347 | -0.2176 | No | ||
| 32 | ELL | 9004 | 0.343 | -0.2164 | No | ||
| 33 | GTF2H3 | 9134 | 0.315 | -0.2213 | No | ||
| 34 | CDK9 | 10456 | 0.030 | -0.2925 | No | ||
| 35 | PAPOLA | 10461 | 0.030 | -0.2925 | No | ||
| 36 | POLR2A | 10546 | 0.010 | -0.2970 | No | ||
| 37 | SF4 | 11174 | -0.134 | -0.3300 | No | ||
| 38 | SFRS9 | 11352 | -0.172 | -0.3384 | No | ||
| 39 | TAF4 | 11511 | -0.211 | -0.3456 | No | ||
| 40 | SNRPB2 | 11731 | -0.260 | -0.3558 | No | ||
| 41 | PCBP1 | 11894 | -0.298 | -0.3626 | No | ||
| 42 | GTF2F1 | 12759 | -0.510 | -0.4060 | No | ||
| 43 | PTBP1 | 12809 | -0.523 | -0.4052 | No | ||
| 44 | FUS | 12866 | -0.537 | -0.4048 | No | ||
| 45 | TCEB3 | 12896 | -0.544 | -0.4029 | No | ||
| 46 | MAGOH | 12985 | -0.571 | -0.4039 | No | ||
| 47 | THOC4 | 13068 | -0.594 | -0.4045 | No | ||
| 48 | SNRPG | 13132 | -0.612 | -0.4040 | No | ||
| 49 | TAF9 | 13299 | -0.660 | -0.4087 | No | ||
| 50 | PCF11 | 14001 | -0.866 | -0.4410 | No | ||
| 51 | SNRPF | 14374 | -0.989 | -0.4547 | No | ||
| 52 | PCBP2 | 14576 | -1.051 | -0.4588 | Yes | ||
| 53 | ERCC3 | 14676 | -1.088 | -0.4571 | Yes | ||
| 54 | GTF2A1 | 14698 | -1.099 | -0.4512 | Yes | ||
| 55 | EFTUD2 | 14861 | -1.156 | -0.4525 | Yes | ||
| 56 | GTF2F2 | 15008 | -1.211 | -0.4525 | Yes | ||
| 57 | SNRPD3 | 15028 | -1.216 | -0.4457 | Yes | ||
| 58 | SF3A1 | 15078 | -1.237 | -0.4404 | Yes | ||
| 59 | YBX1 | 15207 | -1.289 | -0.4390 | Yes | ||
| 60 | SFRS7 | 15210 | -1.289 | -0.4308 | Yes | ||
| 61 | CSTF1 | 15269 | -1.315 | -0.4255 | Yes | ||
| 62 | MNAT1 | 15294 | -1.324 | -0.4182 | Yes | ||
| 63 | METTL3 | 15387 | -1.359 | -0.4144 | Yes | ||
| 64 | DHX9 | 15521 | -1.413 | -0.4125 | Yes | ||
| 65 | POLR2C | 15650 | -1.466 | -0.4100 | Yes | ||
| 66 | SF3B2 | 15670 | -1.478 | -0.4015 | Yes | ||
| 67 | SNRPA1 | 15681 | -1.483 | -0.3924 | Yes | ||
| 68 | DNAJC8 | 15832 | -1.547 | -0.3906 | Yes | ||
| 69 | NCBP2 | 15840 | -1.552 | -0.3809 | Yes | ||
| 70 | SFRS5 | 15877 | -1.569 | -0.3728 | Yes | ||
| 71 | TAF11 | 15919 | -1.587 | -0.3648 | Yes | ||
| 72 | PRPF8 | 16092 | -1.668 | -0.3633 | Yes | ||
| 73 | SF3B3 | 16119 | -1.680 | -0.3539 | Yes | ||
| 74 | CCNT2 | 16150 | -1.693 | -0.3446 | Yes | ||
| 75 | CPSF1 | 16348 | -1.786 | -0.3437 | Yes | ||
| 76 | RBM5 | 16365 | -1.792 | -0.3330 | Yes | ||
| 77 | PRPF4 | 16493 | -1.852 | -0.3279 | Yes | ||
| 78 | TBP | 16552 | -1.880 | -0.3189 | Yes | ||
| 79 | RNMT | 17000 | -2.120 | -0.3294 | Yes | ||
| 80 | PABPN1 | 17007 | -2.123 | -0.3161 | Yes | ||
| 81 | RBMX | 17023 | -2.135 | -0.3031 | Yes | ||
| 82 | PHF5A | 17060 | -2.150 | -0.2912 | Yes | ||
| 83 | SF3A3 | 17334 | -2.311 | -0.2910 | Yes | ||
| 84 | SFRS2 | 17548 | -2.439 | -0.2868 | Yes | ||
| 85 | CCNT1 | 17639 | -2.498 | -0.2756 | Yes | ||
| 86 | GTF2H1 | 17644 | -2.502 | -0.2597 | Yes | ||
| 87 | GTF2E2 | 17658 | -2.510 | -0.2442 | Yes | ||
| 88 | RNGTT | 17757 | -2.575 | -0.2329 | Yes | ||
| 89 | GTF2H2 | 17799 | -2.602 | -0.2183 | Yes | ||
| 90 | SFRS3 | 17871 | -2.665 | -0.2050 | Yes | ||
| 91 | GTF2E1 | 17912 | -2.692 | -0.1898 | Yes | ||
| 92 | TCEB1 | 18070 | -2.831 | -0.1800 | Yes | ||
| 93 | CSTF3 | 18086 | -2.844 | -0.1625 | Yes | ||
| 94 | TAF13 | 18182 | -2.948 | -0.1486 | Yes | ||
| 95 | CCAR1 | 18251 | -3.060 | -0.1325 | Yes | ||
| 96 | CCNH | 18320 | -3.158 | -0.1159 | Yes | ||
| 97 | SFRS1 | 18333 | -3.183 | -0.0960 | Yes | ||
| 98 | CSTF2 | 18338 | -3.191 | -0.0756 | Yes | ||
| 99 | SMC1A | 18339 | -3.191 | -0.0551 | Yes | ||
| 100 | SFRS6 | 18350 | -3.206 | -0.0349 | Yes | ||
| 101 | GTF2B | 18495 | -3.569 | -0.0197 | Yes | ||
| 102 | TAF5 | 18566 | -4.062 | 0.0027 | Yes |
