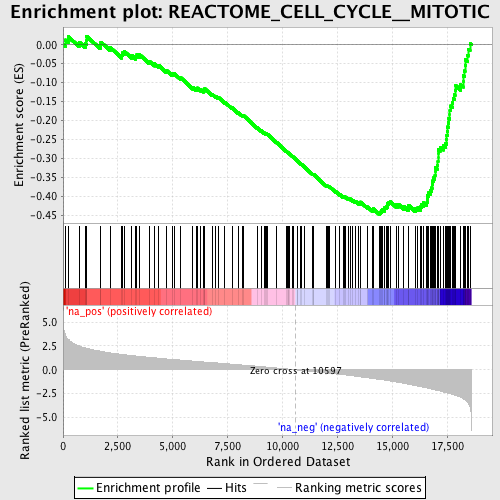
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | -0.44727105 |
| Normalized Enrichment Score (NES) | -2.1587412 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00276483 |
| FWER p-Value | 0.021 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDK4 | 103 | 3.623 | 0.0126 | No | ||
| 2 | CSNK1E | 239 | 3.173 | 0.0212 | No | ||
| 3 | TK2 | 732 | 2.480 | 0.0070 | No | ||
| 4 | TUBB4 | 1007 | 2.279 | 0.0036 | No | ||
| 5 | TUBA1A | 1063 | 2.244 | 0.0119 | No | ||
| 6 | DCTN2 | 1071 | 2.241 | 0.0227 | No | ||
| 7 | PSMD9 | 1691 | 1.948 | -0.0010 | No | ||
| 8 | RPA2 | 1719 | 1.935 | 0.0072 | No | ||
| 9 | CCNE1 | 2145 | 1.770 | -0.0069 | No | ||
| 10 | NEK2 | 2671 | 1.615 | -0.0272 | No | ||
| 11 | PSMB2 | 2714 | 1.601 | -0.0215 | No | ||
| 12 | CDC25B | 2787 | 1.580 | -0.0175 | No | ||
| 13 | CENPE | 3121 | 1.491 | -0.0280 | No | ||
| 14 | PLK1 | 3309 | 1.442 | -0.0309 | No | ||
| 15 | PMF1 | 3364 | 1.430 | -0.0266 | No | ||
| 16 | LIG1 | 3491 | 1.400 | -0.0265 | No | ||
| 17 | CDC20 | 3939 | 1.299 | -0.0441 | No | ||
| 18 | PSMB10 | 4161 | 1.254 | -0.0498 | No | ||
| 19 | PRKACA | 4367 | 1.207 | -0.0549 | No | ||
| 20 | MCM7 | 4727 | 1.130 | -0.0686 | No | ||
| 21 | RFC5 | 4981 | 1.084 | -0.0769 | No | ||
| 22 | PRKAR2B | 5066 | 1.069 | -0.0761 | No | ||
| 23 | PSMC4 | 5367 | 1.009 | -0.0872 | No | ||
| 24 | PSMA7 | 5916 | 0.906 | -0.1124 | No | ||
| 25 | PSMD5 | 6081 | 0.877 | -0.1168 | No | ||
| 26 | TYMS | 6108 | 0.872 | -0.1139 | No | ||
| 27 | POLD1 | 6261 | 0.843 | -0.1179 | No | ||
| 28 | KIF20A | 6391 | 0.822 | -0.1207 | No | ||
| 29 | SSNA1 | 6427 | 0.814 | -0.1186 | No | ||
| 30 | CDK7 | 6460 | 0.809 | -0.1162 | No | ||
| 31 | PSMC1 | 6801 | 0.743 | -0.1309 | No | ||
| 32 | POLE2 | 6949 | 0.721 | -0.1353 | No | ||
| 33 | NUMA1 | 7085 | 0.700 | -0.1390 | No | ||
| 34 | ACTR1A | 7374 | 0.650 | -0.1514 | No | ||
| 35 | POLD2 | 7698 | 0.587 | -0.1659 | No | ||
| 36 | BUB3 | 7978 | 0.535 | -0.1783 | No | ||
| 37 | PSMA5 | 8161 | 0.498 | -0.1857 | No | ||
| 38 | CLIP1 | 8232 | 0.487 | -0.1870 | No | ||
| 39 | POLA1 | 8844 | 0.374 | -0.2182 | No | ||
| 40 | MCM5 | 9024 | 0.339 | -0.2262 | No | ||
| 41 | TUBA4A | 9166 | 0.306 | -0.2323 | No | ||
| 42 | MCM2 | 9236 | 0.291 | -0.2346 | No | ||
| 43 | E2F1 | 9255 | 0.288 | -0.2341 | No | ||
| 44 | POLA2 | 9324 | 0.275 | -0.2364 | No | ||
| 45 | PSMB4 | 9741 | 0.181 | -0.2581 | No | ||
| 46 | BUB1 | 10189 | 0.087 | -0.2818 | No | ||
| 47 | MCM3 | 10237 | 0.078 | -0.2840 | No | ||
| 48 | PSMB9 | 10277 | 0.070 | -0.2857 | No | ||
| 49 | PSMB5 | 10292 | 0.067 | -0.2861 | No | ||
| 50 | PSMA6 | 10299 | 0.065 | -0.2861 | No | ||
| 51 | RFC3 | 10437 | 0.034 | -0.2934 | No | ||
| 52 | DYNLL1 | 10492 | 0.022 | -0.2962 | No | ||
| 53 | UBE2C | 10662 | -0.017 | -0.3053 | No | ||
| 54 | CCNA1 | 10686 | -0.025 | -0.3064 | No | ||
| 55 | PSMD8 | 10838 | -0.059 | -0.3143 | No | ||
| 56 | XPO1 | 10884 | -0.068 | -0.3164 | No | ||
| 57 | PSMC3 | 10997 | -0.093 | -0.3220 | No | ||
| 58 | DCTN3 | 11376 | -0.178 | -0.3415 | No | ||
| 59 | CLASP2 | 11399 | -0.184 | -0.3418 | No | ||
| 60 | UBE2D1 | 11408 | -0.187 | -0.3413 | No | ||
| 61 | PTTG1 | 12020 | -0.329 | -0.3727 | No | ||
| 62 | PSMB1 | 12039 | -0.334 | -0.3720 | No | ||
| 63 | MCM10 | 12091 | -0.345 | -0.3730 | No | ||
| 64 | TUBG1 | 12162 | -0.363 | -0.3750 | No | ||
| 65 | SDCCAG8 | 12421 | -0.425 | -0.3868 | No | ||
| 66 | CENPC1 | 12604 | -0.474 | -0.3943 | No | ||
| 67 | YWHAE | 12773 | -0.513 | -0.4008 | No | ||
| 68 | CDKN1A | 12803 | -0.522 | -0.3998 | No | ||
| 69 | DHFR | 12879 | -0.540 | -0.4011 | No | ||
| 70 | CDC25A | 13018 | -0.579 | -0.4057 | No | ||
| 71 | SEC13 | 13084 | -0.598 | -0.4062 | No | ||
| 72 | FEN1 | 13207 | -0.630 | -0.4097 | No | ||
| 73 | ORC2L | 13315 | -0.665 | -0.4121 | No | ||
| 74 | CLASP1 | 13461 | -0.707 | -0.4164 | No | ||
| 75 | PSMD2 | 13539 | -0.730 | -0.4169 | No | ||
| 76 | PSME3 | 13560 | -0.736 | -0.4143 | No | ||
| 77 | CENPA | 13875 | -0.829 | -0.4272 | No | ||
| 78 | NUP43 | 14122 | -0.908 | -0.4359 | No | ||
| 79 | BIRC5 | 14134 | -0.913 | -0.4320 | No | ||
| 80 | CCNB2 | 14418 | -1.002 | -0.4422 | Yes | ||
| 81 | PSMC5 | 14485 | -1.021 | -0.4407 | Yes | ||
| 82 | ORC1L | 14523 | -1.032 | -0.4375 | Yes | ||
| 83 | PSMD11 | 14541 | -1.037 | -0.4332 | Yes | ||
| 84 | PPP1CC | 14643 | -1.074 | -0.4333 | Yes | ||
| 85 | PSMA1 | 14663 | -1.082 | -0.4289 | Yes | ||
| 86 | RANGAP1 | 14734 | -1.109 | -0.4272 | Yes | ||
| 87 | CDC16 | 14779 | -1.128 | -0.4239 | Yes | ||
| 88 | WEE1 | 14784 | -1.131 | -0.4184 | Yes | ||
| 89 | KNTC1 | 14846 | -1.150 | -0.4160 | Yes | ||
| 90 | RFC4 | 14912 | -1.173 | -0.4136 | Yes | ||
| 91 | PSMC6 | 15180 | -1.279 | -0.4216 | Yes | ||
| 92 | MNAT1 | 15294 | -1.324 | -0.4211 | Yes | ||
| 93 | KIF2A | 15533 | -1.417 | -0.4269 | Yes | ||
| 94 | GMNN | 15735 | -1.506 | -0.4302 | Yes | ||
| 95 | UBE2E1 | 15751 | -1.513 | -0.4235 | Yes | ||
| 96 | RPA3 | 16062 | -1.655 | -0.4319 | Yes | ||
| 97 | ZWINT | 16172 | -1.700 | -0.4293 | Yes | ||
| 98 | CDC25C | 16298 | -1.762 | -0.4272 | Yes | ||
| 99 | ORC5L | 16341 | -1.784 | -0.4206 | Yes | ||
| 100 | STAG2 | 16433 | -1.825 | -0.4164 | Yes | ||
| 101 | CDC45L | 16582 | -1.897 | -0.4149 | Yes | ||
| 102 | SGOL1 | 16597 | -1.909 | -0.4060 | Yes | ||
| 103 | CETN2 | 16611 | -1.917 | -0.3971 | Yes | ||
| 104 | POLD3 | 16664 | -1.938 | -0.3902 | Yes | ||
| 105 | YWHAG | 16732 | -1.980 | -0.3839 | Yes | ||
| 106 | NUP37 | 16797 | -2.018 | -0.3773 | Yes | ||
| 107 | ZW10 | 16818 | -2.028 | -0.3682 | Yes | ||
| 108 | CUL1 | 16836 | -2.035 | -0.3589 | Yes | ||
| 109 | PSMD3 | 16879 | -2.057 | -0.3509 | Yes | ||
| 110 | SGOL2 | 16947 | -2.091 | -0.3440 | Yes | ||
| 111 | NUP160 | 16956 | -2.095 | -0.3339 | Yes | ||
| 112 | PRIM1 | 16962 | -2.096 | -0.3237 | Yes | ||
| 113 | RANBP2 | 17048 | -2.144 | -0.3175 | Yes | ||
| 114 | PAFAH1B1 | 17084 | -2.165 | -0.3086 | Yes | ||
| 115 | RAD21 | 17089 | -2.167 | -0.2979 | Yes | ||
| 116 | PCNA | 17091 | -2.167 | -0.2871 | Yes | ||
| 117 | MAPRE1 | 17095 | -2.168 | -0.2764 | Yes | ||
| 118 | RPA1 | 17181 | -2.221 | -0.2699 | Yes | ||
| 119 | RB1 | 17323 | -2.301 | -0.2660 | Yes | ||
| 120 | OFD1 | 17431 | -2.367 | -0.2599 | Yes | ||
| 121 | PSMD4 | 17477 | -2.395 | -0.2503 | Yes | ||
| 122 | CDK2 | 17488 | -2.401 | -0.2388 | Yes | ||
| 123 | KIF23 | 17511 | -2.417 | -0.2279 | Yes | ||
| 124 | PSMD7 | 17529 | -2.431 | -0.2166 | Yes | ||
| 125 | CCNB1 | 17554 | -2.442 | -0.2057 | Yes | ||
| 126 | POLE | 17578 | -2.455 | -0.1946 | Yes | ||
| 127 | FGFR1OP | 17627 | -2.490 | -0.1847 | Yes | ||
| 128 | RFC1 | 17631 | -2.492 | -0.1724 | Yes | ||
| 129 | PSMA4 | 17660 | -2.511 | -0.1613 | Yes | ||
| 130 | PLK4 | 17746 | -2.570 | -0.1530 | Yes | ||
| 131 | PSMA2 | 17770 | -2.581 | -0.1413 | Yes | ||
| 132 | BUB1B | 17842 | -2.637 | -0.1320 | Yes | ||
| 133 | PSMA3 | 17865 | -2.659 | -0.1198 | Yes | ||
| 134 | MAD2L1 | 17898 | -2.685 | -0.1081 | Yes | ||
| 135 | NUP107 | 18117 | -2.875 | -0.1055 | Yes | ||
| 136 | CCNA2 | 18230 | -3.023 | -0.0964 | Yes | ||
| 137 | MCM4 | 18238 | -3.033 | -0.0815 | Yes | ||
| 138 | PSMD14 | 18296 | -3.126 | -0.0689 | Yes | ||
| 139 | CCNH | 18320 | -3.158 | -0.0544 | Yes | ||
| 140 | SMC1A | 18339 | -3.191 | -0.0393 | Yes | ||
| 141 | PSMD12 | 18428 | -3.388 | -0.0271 | Yes | ||
| 142 | NDC80 | 18478 | -3.521 | -0.0121 | Yes | ||
| 143 | ORC4L | 18548 | -3.893 | 0.0037 | Yes |
