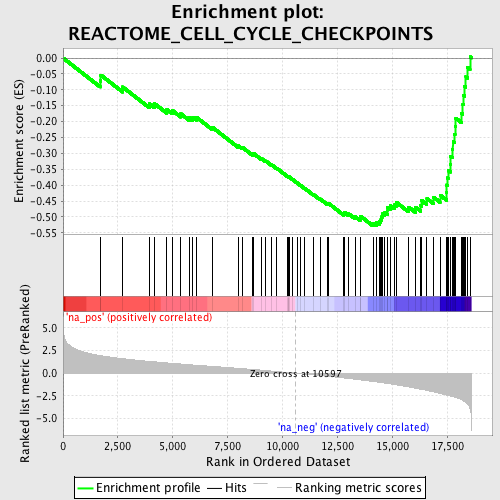
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | -0.5285112 |
| Normalized Enrichment Score (NES) | -2.2570477 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006630839 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMD9 | 1691 | 1.948 | -0.0716 | No | ||
| 2 | RPA2 | 1719 | 1.935 | -0.0535 | No | ||
| 3 | PSMB2 | 2714 | 1.601 | -0.0910 | No | ||
| 4 | CDC20 | 3939 | 1.299 | -0.1440 | No | ||
| 5 | PSMB10 | 4161 | 1.254 | -0.1432 | No | ||
| 6 | MCM7 | 4727 | 1.130 | -0.1623 | No | ||
| 7 | RFC5 | 4981 | 1.084 | -0.1650 | No | ||
| 8 | PSMC4 | 5367 | 1.009 | -0.1757 | No | ||
| 9 | PSMB6 | 5748 | 0.939 | -0.1867 | No | ||
| 10 | PSMA7 | 5916 | 0.906 | -0.1866 | No | ||
| 11 | PSMD5 | 6081 | 0.877 | -0.1866 | No | ||
| 12 | PSMC1 | 6801 | 0.743 | -0.2179 | No | ||
| 13 | BUB3 | 7978 | 0.535 | -0.2759 | No | ||
| 14 | PSMA5 | 8161 | 0.498 | -0.2807 | No | ||
| 15 | RAD1 | 8641 | 0.409 | -0.3024 | No | ||
| 16 | PSMB3 | 8664 | 0.405 | -0.2995 | No | ||
| 17 | MCM5 | 9024 | 0.339 | -0.3155 | No | ||
| 18 | MCM2 | 9236 | 0.291 | -0.3239 | No | ||
| 19 | TP53 | 9507 | 0.232 | -0.3362 | No | ||
| 20 | PSMB4 | 9741 | 0.181 | -0.3469 | No | ||
| 21 | MCM3 | 10237 | 0.078 | -0.3728 | No | ||
| 22 | PSMB9 | 10277 | 0.070 | -0.3742 | No | ||
| 23 | PSMB5 | 10292 | 0.067 | -0.3743 | No | ||
| 24 | PSMA6 | 10299 | 0.065 | -0.3740 | No | ||
| 25 | RFC3 | 10437 | 0.034 | -0.3810 | No | ||
| 26 | UBE2C | 10662 | -0.017 | -0.3929 | No | ||
| 27 | PSMD8 | 10838 | -0.059 | -0.4018 | No | ||
| 28 | PSMC3 | 10997 | -0.093 | -0.4093 | No | ||
| 29 | UBE2D1 | 11408 | -0.187 | -0.4296 | No | ||
| 30 | PSMC2 | 11717 | -0.255 | -0.4436 | No | ||
| 31 | PSMB1 | 12039 | -0.334 | -0.4576 | No | ||
| 32 | MCM10 | 12091 | -0.345 | -0.4568 | No | ||
| 33 | CHEK1 | 12792 | -0.517 | -0.4894 | No | ||
| 34 | CDKN1A | 12803 | -0.522 | -0.4847 | No | ||
| 35 | CDC25A | 13018 | -0.579 | -0.4904 | No | ||
| 36 | ORC2L | 13315 | -0.665 | -0.4996 | No | ||
| 37 | PSMD2 | 13539 | -0.730 | -0.5043 | No | ||
| 38 | PSME3 | 13560 | -0.736 | -0.4980 | No | ||
| 39 | ATM | 14127 | -0.911 | -0.5193 | Yes | ||
| 40 | PSME1 | 14286 | -0.960 | -0.5182 | Yes | ||
| 41 | CCNB2 | 14418 | -1.002 | -0.5151 | Yes | ||
| 42 | PSMC5 | 14485 | -1.021 | -0.5084 | Yes | ||
| 43 | ORC1L | 14523 | -1.032 | -0.5000 | Yes | ||
| 44 | PSMD11 | 14541 | -1.037 | -0.4905 | Yes | ||
| 45 | PSMA1 | 14663 | -1.082 | -0.4861 | Yes | ||
| 46 | CDC16 | 14779 | -1.128 | -0.4810 | Yes | ||
| 47 | WEE1 | 14784 | -1.131 | -0.4698 | Yes | ||
| 48 | RFC4 | 14912 | -1.173 | -0.4648 | Yes | ||
| 49 | MCM6 | 15098 | -1.246 | -0.4622 | Yes | ||
| 50 | PSMC6 | 15180 | -1.279 | -0.4537 | Yes | ||
| 51 | UBE2E1 | 15751 | -1.513 | -0.4692 | Yes | ||
| 52 | RPA3 | 16062 | -1.655 | -0.4693 | Yes | ||
| 53 | CDC25C | 16298 | -1.762 | -0.4642 | Yes | ||
| 54 | ORC5L | 16341 | -1.784 | -0.4485 | Yes | ||
| 55 | CDC45L | 16582 | -1.897 | -0.4423 | Yes | ||
| 56 | PSMD3 | 16879 | -2.057 | -0.4376 | Yes | ||
| 57 | RPA1 | 17181 | -2.221 | -0.4314 | Yes | ||
| 58 | PSMD4 | 17477 | -2.395 | -0.4232 | Yes | ||
| 59 | CDK2 | 17488 | -2.401 | -0.3996 | Yes | ||
| 60 | PSMD7 | 17529 | -2.431 | -0.3772 | Yes | ||
| 61 | CCNB1 | 17554 | -2.442 | -0.3539 | Yes | ||
| 62 | RAD9B | 17646 | -2.503 | -0.3336 | Yes | ||
| 63 | PSMA4 | 17660 | -2.511 | -0.3090 | Yes | ||
| 64 | PSMD10 | 17742 | -2.569 | -0.2875 | Yes | ||
| 65 | PSMA2 | 17770 | -2.581 | -0.2630 | Yes | ||
| 66 | BUB1B | 17842 | -2.637 | -0.2403 | Yes | ||
| 67 | PSMA3 | 17865 | -2.659 | -0.2147 | Yes | ||
| 68 | MAD2L1 | 17898 | -2.685 | -0.1893 | Yes | ||
| 69 | CDC7 | 18162 | -2.928 | -0.1740 | Yes | ||
| 70 | HUS1 | 18218 | -3.005 | -0.1467 | Yes | ||
| 71 | MCM4 | 18238 | -3.033 | -0.1172 | Yes | ||
| 72 | PSMD14 | 18296 | -3.126 | -0.0888 | Yes | ||
| 73 | MDM2 | 18357 | -3.228 | -0.0595 | Yes | ||
| 74 | PSMD12 | 18428 | -3.388 | -0.0291 | Yes | ||
| 75 | ORC4L | 18548 | -3.893 | 0.0037 | Yes |
