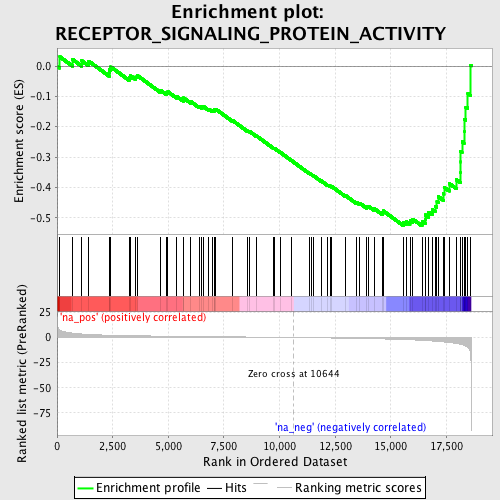
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RECEPTOR_SIGNALING_PROTEIN_ACTIVITY |
| Enrichment Score (ES) | -0.52714515 |
| Normalized Enrichment Score (NES) | -1.7534277 |
| Nominal p-value | 0.0014367816 |
| FDR q-value | 0.33274335 |
| FWER p-Value | 0.984 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TBL3 | 125 | 7.260 | 0.0302 | No | ||
| 2 | SP110 | 698 | 4.350 | 0.0214 | No | ||
| 3 | CSPG4 | 1108 | 3.512 | 0.0172 | No | ||
| 4 | TYRO3 | 1415 | 3.079 | 0.0163 | No | ||
| 5 | GNAZ | 2340 | 2.278 | -0.0219 | No | ||
| 6 | MAP3K10 | 2364 | 2.261 | -0.0117 | No | ||
| 7 | SHC1 | 2417 | 2.229 | -0.0031 | No | ||
| 8 | ACVR1B | 3237 | 1.825 | -0.0380 | No | ||
| 9 | TYROBP | 3303 | 1.796 | -0.0324 | No | ||
| 10 | IRAK1 | 3536 | 1.715 | -0.0362 | No | ||
| 11 | ERBB2 | 3597 | 1.691 | -0.0309 | No | ||
| 12 | MAP4K1 | 4652 | 1.388 | -0.0806 | No | ||
| 13 | MAP3K9 | 4907 | 1.321 | -0.0876 | No | ||
| 14 | IL1F5 | 4980 | 1.302 | -0.0849 | No | ||
| 15 | LYN | 5368 | 1.203 | -0.0997 | No | ||
| 16 | NCR1 | 5668 | 1.128 | -0.1100 | No | ||
| 17 | IFITM1 | 5684 | 1.125 | -0.1051 | No | ||
| 18 | ACVRL1 | 6015 | 1.049 | -0.1176 | No | ||
| 19 | MAP3K6 | 6399 | 0.965 | -0.1334 | No | ||
| 20 | BLNK | 6505 | 0.938 | -0.1343 | No | ||
| 21 | MAPK13 | 6583 | 0.919 | -0.1337 | No | ||
| 22 | MAPK10 | 6822 | 0.864 | -0.1422 | No | ||
| 23 | ACVR2A | 6983 | 0.830 | -0.1466 | No | ||
| 24 | ACVR1C | 7058 | 0.814 | -0.1464 | No | ||
| 25 | MAP3K13 | 7072 | 0.811 | -0.1430 | No | ||
| 26 | CCL4 | 7140 | 0.792 | -0.1426 | No | ||
| 27 | ACVR1 | 7867 | 0.645 | -0.1785 | No | ||
| 28 | MAPK12 | 8571 | 0.491 | -0.2139 | No | ||
| 29 | MAP3K4 | 8643 | 0.476 | -0.2153 | No | ||
| 30 | CD19 | 8961 | 0.408 | -0.2303 | No | ||
| 31 | BAG1 | 9732 | 0.236 | -0.2707 | No | ||
| 32 | SMAD7 | 9750 | 0.233 | -0.2704 | No | ||
| 33 | SMAD6 | 10019 | 0.170 | -0.2840 | No | ||
| 34 | IRAK2 | 10553 | 0.028 | -0.3126 | No | ||
| 35 | IL1RN | 11345 | -0.186 | -0.3543 | No | ||
| 36 | PPARGC1B | 11414 | -0.207 | -0.3569 | No | ||
| 37 | SKIL | 11504 | -0.235 | -0.3605 | No | ||
| 38 | IL18BP | 11864 | -0.340 | -0.3782 | No | ||
| 39 | INSR | 12170 | -0.426 | -0.3924 | No | ||
| 40 | IL1RL1 | 12266 | -0.455 | -0.3953 | No | ||
| 41 | MAPK7 | 12338 | -0.478 | -0.3967 | No | ||
| 42 | KIT | 12949 | -0.677 | -0.4261 | No | ||
| 43 | ALK | 13463 | -0.879 | -0.4493 | No | ||
| 44 | MAP3K5 | 13604 | -0.929 | -0.4521 | No | ||
| 45 | MAP3K7 | 13920 | -1.056 | -0.4638 | No | ||
| 46 | MAPK4 | 13993 | -1.086 | -0.4621 | No | ||
| 47 | IRS1 | 14262 | -1.217 | -0.4704 | No | ||
| 48 | DAXX | 14635 | -1.395 | -0.4834 | No | ||
| 49 | AMHR2 | 14658 | -1.405 | -0.4774 | No | ||
| 50 | MAP3K11 | 15569 | -2.001 | -0.5163 | Yes | ||
| 51 | PLCE1 | 15691 | -2.096 | -0.5122 | Yes | ||
| 52 | ARF1 | 15862 | -2.243 | -0.5100 | Yes | ||
| 53 | ACVR2B | 15985 | -2.360 | -0.5045 | Yes | ||
| 54 | NSMAF | 16405 | -2.799 | -0.5129 | Yes | ||
| 55 | IKBKE | 16547 | -2.984 | -0.5054 | Yes | ||
| 56 | MAPK8 | 16556 | -2.993 | -0.4906 | Yes | ||
| 57 | MAPK14 | 16698 | -3.190 | -0.4820 | Yes | ||
| 58 | TGFBR1 | 16864 | -3.455 | -0.4733 | Yes | ||
| 59 | NLK | 16994 | -3.635 | -0.4618 | Yes | ||
| 60 | ARNT | 17073 | -3.770 | -0.4469 | Yes | ||
| 61 | MAPK1 | 17133 | -3.886 | -0.4303 | Yes | ||
| 62 | CDK5 | 17373 | -4.362 | -0.4210 | Yes | ||
| 63 | MAP3K3 | 17391 | -4.397 | -0.3996 | Yes | ||
| 64 | BAG4 | 17623 | -4.916 | -0.3871 | Yes | ||
| 65 | MAPK9 | 17946 | -5.900 | -0.3745 | Yes | ||
| 66 | MAPK11 | 18122 | -6.671 | -0.3500 | Yes | ||
| 67 | SOCS2 | 18129 | -6.703 | -0.3163 | Yes | ||
| 68 | STAT3 | 18132 | -6.717 | -0.2822 | Yes | ||
| 69 | CD3E | 18203 | -7.143 | -0.2497 | Yes | ||
| 70 | TGFBR2 | 18294 | -7.848 | -0.2147 | Yes | ||
| 71 | STAT1 | 18301 | -7.906 | -0.1748 | Yes | ||
| 72 | STAT2 | 18372 | -8.563 | -0.1351 | Yes | ||
| 73 | CDKN1B | 18463 | -9.688 | -0.0907 | Yes | ||
| 74 | IL4R | 18592 | -19.467 | 0.0013 | Yes |
