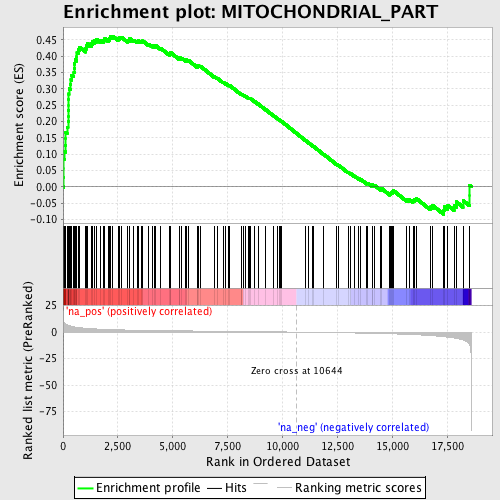
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.46142623 |
| Normalized Enrichment Score (NES) | 1.9571781 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014582851 |
| FWER p-Value | 0.136 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAX | 24 | 10.554 | 0.0284 | Yes | ||
| 2 | PHB2 | 32 | 10.037 | 0.0562 | Yes | ||
| 3 | TIMM50 | 34 | 9.946 | 0.0841 | Yes | ||
| 4 | ABCB6 | 49 | 9.174 | 0.1091 | Yes | ||
| 5 | TIMM13 | 105 | 7.518 | 0.1272 | Yes | ||
| 6 | MRPS10 | 116 | 7.370 | 0.1474 | Yes | ||
| 7 | MRPS15 | 121 | 7.306 | 0.1677 | Yes | ||
| 8 | MRPS18A | 184 | 6.684 | 0.1831 | Yes | ||
| 9 | ATP5E | 223 | 6.409 | 0.1991 | Yes | ||
| 10 | ATP5G1 | 233 | 6.288 | 0.2163 | Yes | ||
| 11 | BCS1L | 239 | 6.248 | 0.2335 | Yes | ||
| 12 | TIMM10 | 253 | 6.177 | 0.2502 | Yes | ||
| 13 | MRPL55 | 256 | 6.167 | 0.2674 | Yes | ||
| 14 | ATP5G2 | 263 | 6.127 | 0.2843 | Yes | ||
| 15 | MRPL12 | 269 | 6.106 | 0.3012 | Yes | ||
| 16 | ETFB | 334 | 5.674 | 0.3137 | Yes | ||
| 17 | NDUFV1 | 357 | 5.602 | 0.3282 | Yes | ||
| 18 | MRPS28 | 387 | 5.471 | 0.3420 | Yes | ||
| 19 | HTRA2 | 493 | 5.037 | 0.3505 | Yes | ||
| 20 | SLC25A1 | 507 | 4.950 | 0.3637 | Yes | ||
| 21 | SLC25A3 | 518 | 4.917 | 0.3770 | Yes | ||
| 22 | UQCRC1 | 541 | 4.840 | 0.3894 | Yes | ||
| 23 | MRPS11 | 625 | 4.545 | 0.3977 | Yes | ||
| 24 | TOMM22 | 631 | 4.523 | 0.4101 | Yes | ||
| 25 | AIFM2 | 686 | 4.386 | 0.4195 | Yes | ||
| 26 | ATP5D | 750 | 4.224 | 0.4280 | Yes | ||
| 27 | ABCF2 | 1034 | 3.636 | 0.4229 | Yes | ||
| 28 | PITRM1 | 1057 | 3.596 | 0.4318 | Yes | ||
| 29 | MRPL10 | 1103 | 3.518 | 0.4392 | Yes | ||
| 30 | NDUFS8 | 1292 | 3.237 | 0.4382 | Yes | ||
| 31 | NDUFS2 | 1334 | 3.172 | 0.4448 | Yes | ||
| 32 | BCKDK | 1427 | 3.058 | 0.4485 | Yes | ||
| 33 | RAB11FIP5 | 1528 | 2.951 | 0.4513 | Yes | ||
| 34 | MRPS12 | 1719 | 2.761 | 0.4488 | Yes | ||
| 35 | IMMT | 1858 | 2.628 | 0.4487 | Yes | ||
| 36 | PPOX | 1902 | 2.586 | 0.4537 | Yes | ||
| 37 | MRPS35 | 2074 | 2.453 | 0.4513 | Yes | ||
| 38 | SUPV3L1 | 2125 | 2.409 | 0.4554 | Yes | ||
| 39 | ATP5O | 2159 | 2.387 | 0.4603 | Yes | ||
| 40 | OPA1 | 2260 | 2.322 | 0.4614 | Yes | ||
| 41 | NDUFS7 | 2507 | 2.176 | 0.4542 | No | ||
| 42 | MRPS24 | 2582 | 2.139 | 0.4562 | No | ||
| 43 | VDAC2 | 2650 | 2.103 | 0.4585 | No | ||
| 44 | MRPS16 | 2939 | 1.950 | 0.4484 | No | ||
| 45 | GRPEL1 | 3027 | 1.915 | 0.4491 | No | ||
| 46 | MAOB | 3037 | 1.911 | 0.4540 | No | ||
| 47 | ATP5F1 | 3220 | 1.832 | 0.4493 | No | ||
| 48 | VDAC3 | 3382 | 1.766 | 0.4455 | No | ||
| 49 | SDHD | 3430 | 1.750 | 0.4479 | No | ||
| 50 | SURF1 | 3552 | 1.706 | 0.4462 | No | ||
| 51 | DBT | 3628 | 1.680 | 0.4468 | No | ||
| 52 | MRPL23 | 3905 | 1.593 | 0.4364 | No | ||
| 53 | PMPCA | 4061 | 1.551 | 0.4323 | No | ||
| 54 | COX6B2 | 4160 | 1.519 | 0.4313 | No | ||
| 55 | BCKDHA | 4208 | 1.505 | 0.4330 | No | ||
| 56 | TIMM23 | 4432 | 1.443 | 0.4250 | No | ||
| 57 | ATP5J | 4847 | 1.336 | 0.4064 | No | ||
| 58 | HCCS | 4874 | 1.329 | 0.4087 | No | ||
| 59 | NDUFS3 | 4902 | 1.322 | 0.4109 | No | ||
| 60 | ATP5A1 | 5317 | 1.218 | 0.3920 | No | ||
| 61 | HSD3B2 | 5322 | 1.215 | 0.3952 | No | ||
| 62 | TFB2M | 5405 | 1.195 | 0.3941 | No | ||
| 63 | COX15 | 5592 | 1.147 | 0.3872 | No | ||
| 64 | TIMM17A | 5622 | 1.140 | 0.3889 | No | ||
| 65 | CASQ1 | 5707 | 1.120 | 0.3875 | No | ||
| 66 | HSD3B1 | 6119 | 1.026 | 0.3681 | No | ||
| 67 | NDUFA6 | 6132 | 1.024 | 0.3704 | No | ||
| 68 | CENTA2 | 6163 | 1.017 | 0.3716 | No | ||
| 69 | POLG2 | 6247 | 0.999 | 0.3699 | No | ||
| 70 | ALDH4A1 | 6880 | 0.851 | 0.3381 | No | ||
| 71 | MRPL40 | 7020 | 0.822 | 0.3329 | No | ||
| 72 | UCP3 | 7297 | 0.762 | 0.3201 | No | ||
| 73 | MFN2 | 7420 | 0.737 | 0.3156 | No | ||
| 74 | ETFA | 7548 | 0.712 | 0.3107 | No | ||
| 75 | MRPS22 | 7572 | 0.706 | 0.3115 | No | ||
| 76 | CYCS | 8120 | 0.587 | 0.2835 | No | ||
| 77 | MTX2 | 8143 | 0.582 | 0.2840 | No | ||
| 78 | MRPS18C | 8240 | 0.564 | 0.2804 | No | ||
| 79 | TIMM17B | 8332 | 0.544 | 0.2770 | No | ||
| 80 | ABCB7 | 8461 | 0.518 | 0.2715 | No | ||
| 81 | CS | 8493 | 0.510 | 0.2713 | No | ||
| 82 | TIMM9 | 8527 | 0.502 | 0.2709 | No | ||
| 83 | NR3C1 | 8735 | 0.461 | 0.2610 | No | ||
| 84 | BCKDHB | 8887 | 0.425 | 0.2540 | No | ||
| 85 | MRPL52 | 9239 | 0.348 | 0.2360 | No | ||
| 86 | BNIP3 | 9579 | 0.270 | 0.2184 | No | ||
| 87 | GATM | 9757 | 0.232 | 0.2095 | No | ||
| 88 | ATP5G3 | 9871 | 0.206 | 0.2040 | No | ||
| 89 | NDUFA9 | 9896 | 0.199 | 0.2032 | No | ||
| 90 | ATP5C1 | 9944 | 0.189 | 0.2012 | No | ||
| 91 | VDAC1 | 11042 | -0.101 | 0.1422 | No | ||
| 92 | UQCRB | 11202 | -0.146 | 0.1340 | No | ||
| 93 | PHB | 11377 | -0.198 | 0.1251 | No | ||
| 94 | MRPS36 | 11420 | -0.209 | 0.1234 | No | ||
| 95 | NFS1 | 11873 | -0.342 | 0.0999 | No | ||
| 96 | SDHA | 12456 | -0.515 | 0.0699 | No | ||
| 97 | TOMM34 | 12568 | -0.551 | 0.0655 | No | ||
| 98 | PIN4 | 12988 | -0.692 | 0.0447 | No | ||
| 99 | HADHB | 13094 | -0.735 | 0.0411 | No | ||
| 100 | TIMM8B | 13282 | -0.806 | 0.0333 | No | ||
| 101 | NDUFAB1 | 13477 | -0.884 | 0.0253 | No | ||
| 102 | UQCRH | 13569 | -0.915 | 0.0229 | No | ||
| 103 | NDUFA1 | 13837 | -1.024 | 0.0113 | No | ||
| 104 | FIS1 | 13885 | -1.042 | 0.0117 | No | ||
| 105 | NNT | 14083 | -1.130 | 0.0043 | No | ||
| 106 | SLC25A22 | 14088 | -1.133 | 0.0072 | No | ||
| 107 | ACN9 | 14185 | -1.180 | 0.0053 | No | ||
| 108 | MRPL32 | 14480 | -1.320 | -0.0069 | No | ||
| 109 | NDUFA2 | 14505 | -1.328 | -0.0044 | No | ||
| 110 | NDUFS1 | 14892 | -1.539 | -0.0210 | No | ||
| 111 | DNAJA3 | 14938 | -1.568 | -0.0190 | No | ||
| 112 | ATP5B | 14979 | -1.593 | -0.0167 | No | ||
| 113 | MPV17 | 15028 | -1.615 | -0.0147 | No | ||
| 114 | OGDH | 15045 | -1.626 | -0.0110 | No | ||
| 115 | MRPS21 | 15668 | -2.072 | -0.0389 | No | ||
| 116 | MRPL51 | 15782 | -2.171 | -0.0389 | No | ||
| 117 | SLC25A15 | 15948 | -2.321 | -0.0413 | No | ||
| 118 | RAF1 | 16017 | -2.394 | -0.0382 | No | ||
| 119 | NDUFA13 | 16083 | -2.464 | -0.0348 | No | ||
| 120 | TIMM44 | 16747 | -3.252 | -0.0615 | No | ||
| 121 | RHOT1 | 16844 | -3.424 | -0.0571 | No | ||
| 122 | SLC25A11 | 17353 | -4.314 | -0.0725 | No | ||
| 123 | RHOT2 | 17371 | -4.360 | -0.0611 | No | ||
| 124 | NDUFS4 | 17535 | -4.732 | -0.0567 | No | ||
| 125 | ACADM | 17820 | -5.462 | -0.0567 | No | ||
| 126 | ALAS2 | 17923 | -5.841 | -0.0458 | No | ||
| 127 | CASP7 | 18227 | -7.255 | -0.0418 | No | ||
| 128 | MCL1 | 18515 | -11.142 | -0.0260 | No | ||
| 129 | BCL2 | 18517 | -11.183 | 0.0054 | No |