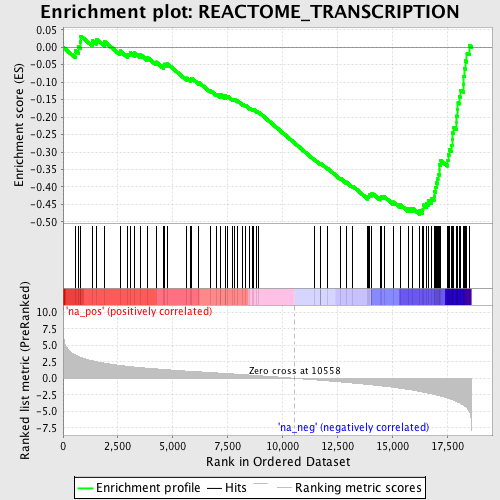
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_TRANSCRIPTION |
| Enrichment Score (ES) | -0.47879797 |
| Normalized Enrichment Score (NES) | -2.0946674 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0052415836 |
| FWER p-Value | 0.03 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPK3 | 561 | 3.505 | -0.0102 | No | ||
| 2 | TCEB2 | 686 | 3.299 | 0.0021 | No | ||
| 3 | TTF1 | 777 | 3.172 | 0.0154 | No | ||
| 4 | TAF6 | 808 | 3.141 | 0.0318 | No | ||
| 5 | TAF12 | 1333 | 2.646 | 0.0187 | No | ||
| 6 | ERCC2 | 1515 | 2.512 | 0.0234 | No | ||
| 7 | TH1L | 1887 | 2.280 | 0.0164 | No | ||
| 8 | POLR2E | 2593 | 1.951 | -0.0104 | No | ||
| 9 | RDBP | 2952 | 1.797 | -0.0194 | No | ||
| 10 | GTF2A2 | 3050 | 1.764 | -0.0145 | No | ||
| 11 | SNAPC2 | 3262 | 1.701 | -0.0162 | No | ||
| 12 | SUPT4H1 | 3517 | 1.622 | -0.0206 | No | ||
| 13 | U2AF2 | 3837 | 1.528 | -0.0290 | No | ||
| 14 | CDK9 | 4233 | 1.427 | -0.0422 | No | ||
| 15 | NFIA | 4585 | 1.334 | -0.0534 | No | ||
| 16 | POLR2A | 4625 | 1.325 | -0.0480 | No | ||
| 17 | TAF1C | 4750 | 1.297 | -0.0472 | No | ||
| 18 | POLR2C | 5622 | 1.092 | -0.0880 | No | ||
| 19 | SFRS9 | 5800 | 1.053 | -0.0915 | No | ||
| 20 | U2AF1 | 5857 | 1.043 | -0.0885 | No | ||
| 21 | NFX1 | 6186 | 0.974 | -0.1006 | No | ||
| 22 | CDK7 | 6702 | 0.871 | -0.1234 | No | ||
| 23 | POLR2J | 7000 | 0.811 | -0.1348 | No | ||
| 24 | GTF3C1 | 7159 | 0.781 | -0.1388 | No | ||
| 25 | SNAPC4 | 7169 | 0.779 | -0.1349 | No | ||
| 26 | PTRF | 7392 | 0.730 | -0.1427 | No | ||
| 27 | ERCC3 | 7393 | 0.729 | -0.1385 | No | ||
| 28 | POLR2G | 7510 | 0.704 | -0.1407 | No | ||
| 29 | POLRMT | 7726 | 0.662 | -0.1485 | No | ||
| 30 | POLR2I | 7807 | 0.645 | -0.1491 | No | ||
| 31 | HIST4H4 | 7951 | 0.613 | -0.1533 | No | ||
| 32 | SSB | 8189 | 0.559 | -0.1629 | No | ||
| 33 | POLR2H | 8318 | 0.530 | -0.1668 | No | ||
| 34 | TAF4 | 8513 | 0.489 | -0.1744 | No | ||
| 35 | NFIB | 8635 | 0.460 | -0.1783 | No | ||
| 36 | SRRM1 | 8679 | 0.449 | -0.1781 | No | ||
| 37 | ELL | 8830 | 0.411 | -0.1838 | No | ||
| 38 | TAF10 | 8914 | 0.391 | -0.1861 | No | ||
| 39 | POLR2B | 11441 | -0.219 | -0.3212 | No | ||
| 40 | POLR3A | 11708 | -0.288 | -0.3339 | No | ||
| 41 | NUDT21 | 11717 | -0.291 | -0.3326 | No | ||
| 42 | GTF2F2 | 11729 | -0.294 | -0.3315 | No | ||
| 43 | CCNT2 | 12065 | -0.387 | -0.3474 | No | ||
| 44 | GTF2F1 | 12629 | -0.540 | -0.3747 | No | ||
| 45 | PAPOLA | 12900 | -0.615 | -0.3857 | No | ||
| 46 | PCF11 | 13200 | -0.708 | -0.3978 | No | ||
| 47 | NFIC | 13892 | -0.922 | -0.4298 | No | ||
| 48 | GTF2H3 | 13926 | -0.936 | -0.4262 | No | ||
| 49 | TAF1B | 13949 | -0.942 | -0.4220 | No | ||
| 50 | RNPS1 | 14036 | -0.969 | -0.4211 | No | ||
| 51 | NCBP2 | 14072 | -0.983 | -0.4173 | No | ||
| 52 | SFRS7 | 14444 | -1.118 | -0.4309 | No | ||
| 53 | POLR3D | 14502 | -1.136 | -0.4275 | No | ||
| 54 | CSTF1 | 14626 | -1.183 | -0.4273 | No | ||
| 55 | RNMT | 15059 | -1.364 | -0.4428 | No | ||
| 56 | CPSF1 | 15369 | -1.506 | -0.4509 | No | ||
| 57 | TAF9 | 15742 | -1.678 | -0.4613 | No | ||
| 58 | UBTF | 15914 | -1.772 | -0.4604 | No | ||
| 59 | MAGOH | 16256 | -1.972 | -0.4675 | Yes | ||
| 60 | GTF2B | 16398 | -2.072 | -0.4632 | Yes | ||
| 61 | SNAPC1 | 16403 | -2.076 | -0.4515 | Yes | ||
| 62 | THOC4 | 16562 | -2.186 | -0.4475 | Yes | ||
| 63 | POU2F1 | 16649 | -2.246 | -0.4392 | Yes | ||
| 64 | GTF2E2 | 16789 | -2.342 | -0.4333 | Yes | ||
| 65 | RNGTT | 16910 | -2.429 | -0.4258 | Yes | ||
| 66 | CSTF3 | 16935 | -2.449 | -0.4131 | Yes | ||
| 67 | TBP | 16974 | -2.479 | -0.4009 | Yes | ||
| 68 | SFRS3 | 17008 | -2.503 | -0.3883 | Yes | ||
| 69 | POLR1A | 17069 | -2.558 | -0.3769 | Yes | ||
| 70 | PABPN1 | 17088 | -2.574 | -0.3631 | Yes | ||
| 71 | TAF11 | 17144 | -2.628 | -0.3510 | Yes | ||
| 72 | CBX3 | 17146 | -2.632 | -0.3359 | Yes | ||
| 73 | TCEB3 | 17196 | -2.681 | -0.3232 | Yes | ||
| 74 | SNAPC5 | 17512 | -2.967 | -0.3231 | Yes | ||
| 75 | CSTF2 | 17550 | -3.014 | -0.3078 | Yes | ||
| 76 | NFIX | 17591 | -3.058 | -0.2924 | Yes | ||
| 77 | CCNH | 17706 | -3.175 | -0.2804 | Yes | ||
| 78 | TCEB1 | 17730 | -3.203 | -0.2632 | Yes | ||
| 79 | SFRS5 | 17737 | -3.212 | -0.2451 | Yes | ||
| 80 | SFRS2 | 17780 | -3.269 | -0.2286 | Yes | ||
| 81 | GTF2E1 | 17933 | -3.496 | -0.2168 | Yes | ||
| 82 | TAF13 | 17936 | -3.499 | -0.1968 | Yes | ||
| 83 | GTF2H1 | 17984 | -3.579 | -0.1788 | Yes | ||
| 84 | TFAM | 17997 | -3.595 | -0.1588 | Yes | ||
| 85 | MNAT1 | 18068 | -3.691 | -0.1414 | Yes | ||
| 86 | GTF2H2 | 18122 | -3.791 | -0.1225 | Yes | ||
| 87 | SFRS6 | 18258 | -4.074 | -0.1064 | Yes | ||
| 88 | CCNT1 | 18265 | -4.084 | -0.0833 | Yes | ||
| 89 | GTF2A1 | 18290 | -4.150 | -0.0608 | Yes | ||
| 90 | TAF5 | 18333 | -4.253 | -0.0386 | Yes | ||
| 91 | SFRS1 | 18407 | -4.486 | -0.0168 | Yes | ||
| 92 | TAF1A | 18506 | -4.887 | 0.0059 | Yes |
