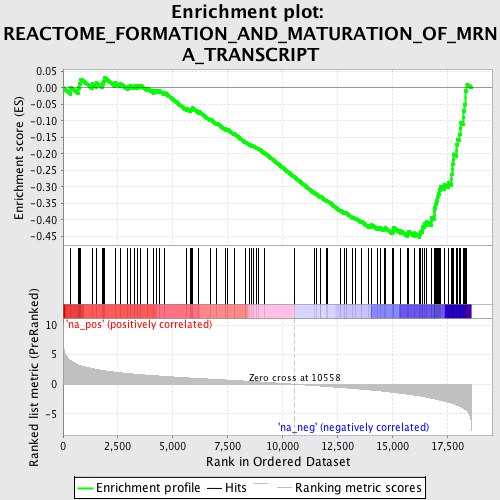
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | -0.45369923 |
| Normalized Enrichment Score (NES) | -2.0238693 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009017695 |
| FWER p-Value | 0.093 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SF3A2 | 348 | 3.973 | 0.0017 | No | ||
| 2 | TCEB2 | 686 | 3.299 | 0.0006 | No | ||
| 3 | CD2BP2 | 752 | 3.211 | 0.0137 | No | ||
| 4 | TAF6 | 808 | 3.141 | 0.0269 | No | ||
| 5 | TAF12 | 1333 | 2.646 | 0.0123 | No | ||
| 6 | ERCC2 | 1515 | 2.512 | 0.0155 | No | ||
| 7 | SF4 | 1784 | 2.340 | 0.0131 | No | ||
| 8 | SNRPA | 1859 | 2.297 | 0.0210 | No | ||
| 9 | TH1L | 1887 | 2.280 | 0.0313 | No | ||
| 10 | SNRPB | 2365 | 2.043 | 0.0161 | No | ||
| 11 | POLR2E | 2593 | 1.951 | 0.0139 | No | ||
| 12 | RDBP | 2952 | 1.797 | 0.0039 | No | ||
| 13 | GTF2A2 | 3050 | 1.764 | 0.0077 | No | ||
| 14 | SNRPE | 3243 | 1.707 | 0.0062 | No | ||
| 15 | SF3B1 | 3399 | 1.661 | 0.0064 | No | ||
| 16 | SUPT4H1 | 3517 | 1.622 | 0.0085 | No | ||
| 17 | U2AF2 | 3837 | 1.528 | -0.0009 | No | ||
| 18 | SF3A1 | 4128 | 1.452 | -0.0090 | No | ||
| 19 | CDK9 | 4233 | 1.427 | -0.0073 | No | ||
| 20 | SNRPD2 | 4380 | 1.387 | -0.0080 | No | ||
| 21 | POLR2A | 4625 | 1.325 | -0.0143 | No | ||
| 22 | POLR2C | 5622 | 1.092 | -0.0625 | No | ||
| 23 | SFRS9 | 5800 | 1.053 | -0.0666 | No | ||
| 24 | U2AF1 | 5857 | 1.043 | -0.0642 | No | ||
| 25 | TXNL4A | 5890 | 1.037 | -0.0606 | No | ||
| 26 | NFX1 | 6186 | 0.974 | -0.0715 | No | ||
| 27 | CDK7 | 6702 | 0.871 | -0.0948 | No | ||
| 28 | POLR2J | 7000 | 0.811 | -0.1067 | No | ||
| 29 | ERCC3 | 7393 | 0.729 | -0.1241 | No | ||
| 30 | POLR2G | 7510 | 0.704 | -0.1267 | No | ||
| 31 | POLR2I | 7807 | 0.645 | -0.1394 | No | ||
| 32 | POLR2H | 8318 | 0.530 | -0.1642 | No | ||
| 33 | TAF4 | 8513 | 0.489 | -0.1721 | No | ||
| 34 | PTBP1 | 8605 | 0.467 | -0.1746 | No | ||
| 35 | SRRM1 | 8679 | 0.449 | -0.1762 | No | ||
| 36 | ELL | 8830 | 0.411 | -0.1822 | No | ||
| 37 | TAF10 | 8914 | 0.391 | -0.1847 | No | ||
| 38 | SF3B4 | 9174 | 0.328 | -0.1970 | No | ||
| 39 | SF3B3 | 10535 | 0.005 | -0.2704 | No | ||
| 40 | POLR2B | 11441 | -0.219 | -0.3182 | No | ||
| 41 | PCBP1 | 11530 | -0.242 | -0.3217 | No | ||
| 42 | NUDT21 | 11717 | -0.291 | -0.3302 | No | ||
| 43 | GTF2F2 | 11729 | -0.294 | -0.3293 | No | ||
| 44 | SNRPA1 | 11988 | -0.365 | -0.3413 | No | ||
| 45 | CCNT2 | 12065 | -0.387 | -0.3434 | No | ||
| 46 | GTF2F1 | 12629 | -0.540 | -0.3710 | No | ||
| 47 | SNRPB2 | 12828 | -0.598 | -0.3786 | No | ||
| 48 | PAPOLA | 12900 | -0.615 | -0.3793 | No | ||
| 49 | PCF11 | 13200 | -0.708 | -0.3918 | No | ||
| 50 | SNRPF | 13326 | -0.748 | -0.3947 | No | ||
| 51 | METTL3 | 13585 | -0.825 | -0.4044 | No | ||
| 52 | GTF2H3 | 13926 | -0.936 | -0.4179 | No | ||
| 53 | RNPS1 | 14036 | -0.969 | -0.4188 | No | ||
| 54 | NCBP2 | 14072 | -0.983 | -0.4156 | No | ||
| 55 | FUS | 14343 | -1.078 | -0.4246 | No | ||
| 56 | SFRS7 | 14444 | -1.118 | -0.4242 | No | ||
| 57 | CSTF1 | 14626 | -1.183 | -0.4279 | No | ||
| 58 | EFTUD2 | 14682 | -1.207 | -0.4246 | No | ||
| 59 | PHF5A | 15018 | -1.345 | -0.4358 | No | ||
| 60 | PCBP2 | 15050 | -1.361 | -0.4304 | No | ||
| 61 | RNMT | 15059 | -1.364 | -0.4238 | No | ||
| 62 | CPSF1 | 15369 | -1.506 | -0.4327 | No | ||
| 63 | PRPF8 | 15690 | -1.651 | -0.4414 | No | ||
| 64 | TAF9 | 15742 | -1.678 | -0.4355 | No | ||
| 65 | SNRPG | 16002 | -1.836 | -0.4400 | No | ||
| 66 | MAGOH | 16256 | -1.972 | -0.4435 | Yes | ||
| 67 | PRPF4 | 16297 | -2.000 | -0.4353 | Yes | ||
| 68 | SNRPD3 | 16367 | -2.054 | -0.4285 | Yes | ||
| 69 | GTF2B | 16398 | -2.072 | -0.4194 | Yes | ||
| 70 | SF3B2 | 16465 | -2.115 | -0.4120 | Yes | ||
| 71 | THOC4 | 16562 | -2.186 | -0.4059 | Yes | ||
| 72 | GTF2E2 | 16789 | -2.342 | -0.4060 | Yes | ||
| 73 | DHX9 | 16794 | -2.348 | -0.3941 | Yes | ||
| 74 | RNGTT | 16910 | -2.429 | -0.3877 | Yes | ||
| 75 | CCAR1 | 16928 | -2.442 | -0.3760 | Yes | ||
| 76 | CSTF3 | 16935 | -2.449 | -0.3637 | Yes | ||
| 77 | TBP | 16974 | -2.479 | -0.3530 | Yes | ||
| 78 | SFRS3 | 17008 | -2.503 | -0.3418 | Yes | ||
| 79 | YBX1 | 17046 | -2.535 | -0.3307 | Yes | ||
| 80 | PABPN1 | 17088 | -2.574 | -0.3196 | Yes | ||
| 81 | TAF11 | 17144 | -2.628 | -0.3090 | Yes | ||
| 82 | TCEB3 | 17196 | -2.681 | -0.2979 | Yes | ||
| 83 | SMC1A | 17365 | -2.831 | -0.2924 | Yes | ||
| 84 | CSTF2 | 17550 | -3.014 | -0.2867 | Yes | ||
| 85 | RBM5 | 17702 | -3.169 | -0.2785 | Yes | ||
| 86 | CCNH | 17706 | -3.175 | -0.2623 | Yes | ||
| 87 | TCEB1 | 17730 | -3.203 | -0.2470 | Yes | ||
| 88 | SFRS5 | 17737 | -3.212 | -0.2307 | Yes | ||
| 89 | SFRS2 | 17780 | -3.269 | -0.2161 | Yes | ||
| 90 | SF3A3 | 17808 | -3.301 | -0.2005 | Yes | ||
| 91 | GTF2E1 | 17933 | -3.496 | -0.1891 | Yes | ||
| 92 | TAF13 | 17936 | -3.499 | -0.1711 | Yes | ||
| 93 | GTF2H1 | 17984 | -3.579 | -0.1552 | Yes | ||
| 94 | MNAT1 | 18068 | -3.691 | -0.1406 | Yes | ||
| 95 | RBMX | 18107 | -3.763 | -0.1232 | Yes | ||
| 96 | GTF2H2 | 18122 | -3.791 | -0.1044 | Yes | ||
| 97 | SFRS6 | 18258 | -4.074 | -0.0906 | Yes | ||
| 98 | CCNT1 | 18265 | -4.084 | -0.0698 | Yes | ||
| 99 | GTF2A1 | 18290 | -4.150 | -0.0497 | Yes | ||
| 100 | DNAJC8 | 18332 | -4.253 | -0.0299 | Yes | ||
| 101 | TAF5 | 18333 | -4.253 | -0.0079 | Yes | ||
| 102 | SFRS1 | 18407 | -4.486 | 0.0113 | Yes |
