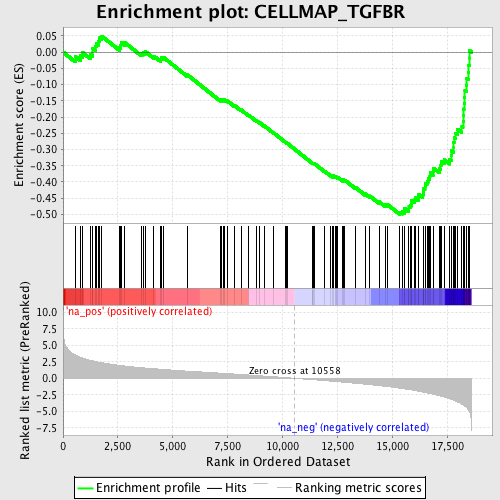
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CELLMAP_TGFBR |
| Enrichment Score (ES) | -0.5001348 |
| Normalized Enrichment Score (NES) | -2.2233968 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ANAPC7 | 555 | 3.516 | -0.0131 | No | ||
| 2 | FZR1 | 784 | 3.162 | -0.0103 | No | ||
| 3 | ARRB2 | 896 | 3.043 | -0.0017 | No | ||
| 4 | AXIN1 | 1247 | 2.717 | -0.0075 | No | ||
| 5 | PIK3R2 | 1340 | 2.638 | 0.0001 | No | ||
| 6 | DAXX | 1341 | 2.637 | 0.0128 | No | ||
| 7 | CAMK2D | 1481 | 2.535 | 0.0174 | No | ||
| 8 | TGFBR2 | 1508 | 2.516 | 0.0281 | No | ||
| 9 | TP53 | 1627 | 2.442 | 0.0334 | No | ||
| 10 | STK11IP | 1651 | 2.427 | 0.0438 | No | ||
| 11 | BTRC | 1757 | 2.357 | 0.0495 | No | ||
| 12 | JUNB | 2580 | 1.958 | 0.0145 | No | ||
| 13 | ANAPC2 | 2626 | 1.934 | 0.0213 | No | ||
| 14 | CDK4 | 2639 | 1.929 | 0.0299 | No | ||
| 15 | VDR | 2799 | 1.865 | 0.0303 | No | ||
| 16 | RUNX2 | 3573 | 1.605 | -0.0038 | No | ||
| 17 | FKBP1A | 3665 | 1.578 | -0.0011 | No | ||
| 18 | SNX6 | 3752 | 1.551 | 0.0017 | No | ||
| 19 | ETS1 | 4141 | 1.449 | -0.0124 | No | ||
| 20 | SMAD3 | 4451 | 1.369 | -0.0225 | No | ||
| 21 | ANAPC10 | 4470 | 1.364 | -0.0169 | No | ||
| 22 | HOXA9 | 4597 | 1.331 | -0.0173 | No | ||
| 23 | MAP3K7IP1 | 5666 | 1.084 | -0.0698 | No | ||
| 24 | ENG | 7164 | 0.779 | -0.1470 | No | ||
| 25 | TGFB1 | 7233 | 0.765 | -0.1470 | No | ||
| 26 | FOXO3 | 7299 | 0.748 | -0.1469 | No | ||
| 27 | SNIP1 | 7369 | 0.735 | -0.1471 | No | ||
| 28 | SDC2 | 7491 | 0.710 | -0.1502 | No | ||
| 29 | SMAD7 | 7830 | 0.638 | -0.1654 | No | ||
| 30 | AR | 8110 | 0.576 | -0.1777 | No | ||
| 31 | CAMK2G | 8451 | 0.501 | -0.1937 | No | ||
| 32 | ZEB2 | 8823 | 0.413 | -0.2118 | No | ||
| 33 | NFYC | 8966 | 0.379 | -0.2176 | No | ||
| 34 | FOS | 9169 | 0.329 | -0.2270 | No | ||
| 35 | SNX1 | 9609 | 0.235 | -0.2495 | No | ||
| 36 | NUP153 | 10151 | 0.107 | -0.2783 | No | ||
| 37 | CAMK2B | 10175 | 0.099 | -0.2790 | No | ||
| 38 | CAV1 | 10226 | 0.085 | -0.2813 | No | ||
| 39 | TGFB3 | 11359 | -0.201 | -0.3415 | No | ||
| 40 | SMAD6 | 11387 | -0.207 | -0.3420 | No | ||
| 41 | XPO1 | 11398 | -0.210 | -0.3415 | No | ||
| 42 | PRKAR1B | 11442 | -0.220 | -0.3428 | No | ||
| 43 | CCNB2 | 11908 | -0.344 | -0.3663 | No | ||
| 44 | FOSB | 12199 | -0.421 | -0.3799 | No | ||
| 45 | CITED1 | 12296 | -0.445 | -0.3830 | No | ||
| 46 | UBE2L3 | 12302 | -0.447 | -0.3811 | No | ||
| 47 | JUN | 12417 | -0.481 | -0.3849 | No | ||
| 48 | CDK6 | 12452 | -0.489 | -0.3844 | No | ||
| 49 | CDKN1A | 12520 | -0.508 | -0.3856 | No | ||
| 50 | SKIL | 12741 | -0.574 | -0.3947 | No | ||
| 51 | ESR1 | 12757 | -0.578 | -0.3928 | No | ||
| 52 | TGFBR3 | 12837 | -0.600 | -0.3942 | No | ||
| 53 | ATF3 | 13344 | -0.753 | -0.4179 | No | ||
| 54 | UBE2D1 | 13769 | -0.885 | -0.4366 | No | ||
| 55 | MYC | 13974 | -0.949 | -0.4430 | No | ||
| 56 | CCND1 | 14405 | -1.103 | -0.4610 | No | ||
| 57 | EID2 | 14677 | -1.204 | -0.4698 | No | ||
| 58 | CCNE1 | 14802 | -1.250 | -0.4705 | No | ||
| 59 | YAP1 | 15351 | -1.497 | -0.4930 | Yes | ||
| 60 | MAP2K6 | 15459 | -1.542 | -0.4913 | Yes | ||
| 61 | COPS5 | 15547 | -1.586 | -0.4884 | Yes | ||
| 62 | ATF2 | 15554 | -1.588 | -0.4811 | Yes | ||
| 63 | SMURF2 | 15755 | -1.683 | -0.4839 | Yes | ||
| 64 | CDC16 | 15763 | -1.686 | -0.4762 | Yes | ||
| 65 | PRKAR2A | 15826 | -1.723 | -0.4712 | Yes | ||
| 66 | ANAPC5 | 15879 | -1.752 | -0.4656 | Yes | ||
| 67 | CUL1 | 15887 | -1.757 | -0.4576 | Yes | ||
| 68 | SNW1 | 16009 | -1.840 | -0.4553 | Yes | ||
| 69 | SNX4 | 16059 | -1.858 | -0.4490 | Yes | ||
| 70 | UBE2D3 | 16195 | -1.941 | -0.4470 | Yes | ||
| 71 | ZFYVE9 | 16208 | -1.951 | -0.4383 | Yes | ||
| 72 | CDC25A | 16402 | -2.075 | -0.4388 | Yes | ||
| 73 | CD44 | 16429 | -2.092 | -0.4302 | Yes | ||
| 74 | UBE2D2 | 16442 | -2.100 | -0.4207 | Yes | ||
| 75 | MAPK14 | 16495 | -2.137 | -0.4133 | Yes | ||
| 76 | TFDP1 | 16526 | -2.165 | -0.4045 | Yes | ||
| 77 | FNTA | 16588 | -2.202 | -0.3973 | Yes | ||
| 78 | PRKCD | 16642 | -2.241 | -0.3894 | Yes | ||
| 79 | DAB2 | 16716 | -2.286 | -0.3823 | Yes | ||
| 80 | HDAC1 | 16722 | -2.288 | -0.3716 | Yes | ||
| 81 | CDC23 | 16863 | -2.396 | -0.3677 | Yes | ||
| 82 | NCOA1 | 16881 | -2.409 | -0.3571 | Yes | ||
| 83 | MAP3K7 | 17162 | -2.642 | -0.3595 | Yes | ||
| 84 | CREBBP | 17218 | -2.699 | -0.3496 | Yes | ||
| 85 | UBE2E1 | 17227 | -2.705 | -0.3370 | Yes | ||
| 86 | TGFBR1 | 17367 | -2.832 | -0.3309 | Yes | ||
| 87 | SNX2 | 17623 | -3.085 | -0.3299 | Yes | ||
| 88 | RB1 | 17687 | -3.152 | -0.3182 | Yes | ||
| 89 | NFYA | 17710 | -3.180 | -0.3041 | Yes | ||
| 90 | RBX1 | 17785 | -3.274 | -0.2924 | Yes | ||
| 91 | SMAD2 | 17789 | -3.278 | -0.2769 | Yes | ||
| 92 | PIAS1 | 17820 | -3.319 | -0.2626 | Yes | ||
| 93 | ZEB1 | 17888 | -3.413 | -0.2498 | Yes | ||
| 94 | HSPA8 | 17987 | -3.581 | -0.2379 | Yes | ||
| 95 | ANAPC4 | 18177 | -3.891 | -0.2295 | Yes | ||
| 96 | RBL1 | 18226 | -3.996 | -0.2129 | Yes | ||
| 97 | CDK2 | 18234 | -4.014 | -0.1940 | Yes | ||
| 98 | KPNB1 | 18242 | -4.033 | -0.1750 | Yes | ||
| 99 | MEF2A | 18273 | -4.111 | -0.1569 | Yes | ||
| 100 | ANAPC1 | 18297 | -4.168 | -0.1382 | Yes | ||
| 101 | RBL2 | 18314 | -4.213 | -0.1188 | Yes | ||
| 102 | SMAD4 | 18375 | -4.376 | -0.1011 | Yes | ||
| 103 | STRAP | 18388 | -4.436 | -0.0805 | Yes | ||
| 104 | CTCF | 18467 | -4.689 | -0.0622 | Yes | ||
| 105 | NFYB | 18488 | -4.777 | -0.0404 | Yes | ||
| 106 | SP1 | 18508 | -4.895 | -0.0179 | Yes | ||
| 107 | MAPK8 | 18511 | -4.936 | 0.0057 | Yes |
