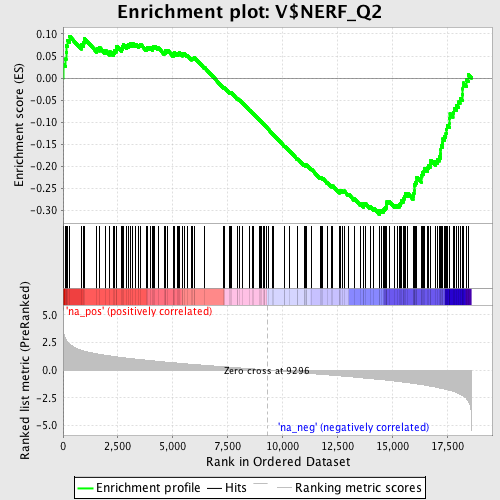
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NERF_Q2 |
| Enrichment Score (ES) | -0.3086141 |
| Normalized Enrichment Score (NES) | -1.4924318 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.47846764 |
| FWER p-Value | 0.993 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FREQ | 1 | 5.316 | 0.0317 | No | ||
| 2 | PCDH7 | 101 | 2.871 | 0.0435 | No | ||
| 3 | ELK3 | 135 | 2.732 | 0.0580 | No | ||
| 4 | FCGR2B | 146 | 2.702 | 0.0736 | No | ||
| 5 | HN1 | 197 | 2.521 | 0.0859 | No | ||
| 6 | SLITRK6 | 288 | 2.347 | 0.0950 | No | ||
| 7 | EXTL2 | 830 | 1.798 | 0.0765 | No | ||
| 8 | STARD8 | 940 | 1.730 | 0.0809 | No | ||
| 9 | ZNF23 | 968 | 1.716 | 0.0897 | No | ||
| 10 | LST1 | 1542 | 1.466 | 0.0673 | No | ||
| 11 | SLCO2B1 | 1657 | 1.421 | 0.0696 | No | ||
| 12 | RPS3 | 1932 | 1.338 | 0.0628 | No | ||
| 13 | VCL | 2133 | 1.276 | 0.0596 | No | ||
| 14 | ELOVL5 | 2300 | 1.236 | 0.0579 | No | ||
| 15 | NDUFS2 | 2339 | 1.227 | 0.0632 | No | ||
| 16 | DIABLO | 2418 | 1.202 | 0.0662 | No | ||
| 17 | STARD13 | 2435 | 1.197 | 0.0724 | No | ||
| 18 | CALM2 | 2679 | 1.139 | 0.0661 | No | ||
| 19 | BCOR | 2691 | 1.135 | 0.0723 | No | ||
| 20 | MYOHD1 | 2740 | 1.122 | 0.0763 | No | ||
| 21 | ARHGAP8 | 2909 | 1.077 | 0.0737 | No | ||
| 22 | TCF12 | 2991 | 1.060 | 0.0756 | No | ||
| 23 | MAP2K6 | 3057 | 1.047 | 0.0783 | No | ||
| 24 | LRRFIP2 | 3172 | 1.023 | 0.0783 | No | ||
| 25 | SHC3 | 3316 | 0.993 | 0.0764 | No | ||
| 26 | FGFR2 | 3452 | 0.964 | 0.0749 | No | ||
| 27 | CDCA3 | 3527 | 0.952 | 0.0766 | No | ||
| 28 | LSAMP | 3802 | 0.894 | 0.0670 | No | ||
| 29 | LOXL3 | 3847 | 0.884 | 0.0699 | No | ||
| 30 | SECISBP2 | 3963 | 0.860 | 0.0688 | No | ||
| 31 | SLC30A7 | 4070 | 0.839 | 0.0681 | No | ||
| 32 | ATOH8 | 4098 | 0.833 | 0.0716 | No | ||
| 33 | GPR132 | 4179 | 0.816 | 0.0721 | No | ||
| 34 | NFATC3 | 4333 | 0.790 | 0.0686 | No | ||
| 35 | NRXN3 | 4602 | 0.737 | 0.0584 | No | ||
| 36 | FKBP10 | 4657 | 0.729 | 0.0599 | No | ||
| 37 | KCND1 | 4669 | 0.727 | 0.0636 | No | ||
| 38 | PAK3 | 4763 | 0.711 | 0.0628 | No | ||
| 39 | TUSC3 | 5017 | 0.665 | 0.0531 | No | ||
| 40 | CD79B | 5023 | 0.664 | 0.0568 | No | ||
| 41 | SH3TC1 | 5064 | 0.659 | 0.0585 | No | ||
| 42 | FURIN | 5206 | 0.633 | 0.0547 | No | ||
| 43 | DPP8 | 5254 | 0.624 | 0.0559 | No | ||
| 44 | TENC1 | 5288 | 0.618 | 0.0578 | No | ||
| 45 | ATP5H | 5434 | 0.591 | 0.0534 | No | ||
| 46 | ITPKB | 5450 | 0.587 | 0.0561 | No | ||
| 47 | PAX6 | 5531 | 0.576 | 0.0552 | No | ||
| 48 | HOXB9 | 5662 | 0.557 | 0.0515 | No | ||
| 49 | GOLGA7 | 5860 | 0.523 | 0.0439 | No | ||
| 50 | TOPORS | 5909 | 0.517 | 0.0444 | No | ||
| 51 | CD79A | 5965 | 0.506 | 0.0445 | No | ||
| 52 | CNTNAP1 | 5967 | 0.506 | 0.0474 | No | ||
| 53 | JAG1 | 6440 | 0.437 | 0.0244 | No | ||
| 54 | KLF13 | 7314 | 0.313 | -0.0210 | No | ||
| 55 | UCHL5 | 7374 | 0.304 | -0.0224 | No | ||
| 56 | ARRB2 | 7604 | 0.270 | -0.0332 | No | ||
| 57 | MMRN2 | 7638 | 0.264 | -0.0334 | No | ||
| 58 | FCER1G | 7652 | 0.263 | -0.0325 | No | ||
| 59 | TRO | 7956 | 0.210 | -0.0477 | No | ||
| 60 | ACTR3 | 7961 | 0.210 | -0.0467 | No | ||
| 61 | CAP1 | 8034 | 0.201 | -0.0494 | No | ||
| 62 | CPNE8 | 8194 | 0.176 | -0.0569 | No | ||
| 63 | CDK5 | 8499 | 0.131 | -0.0726 | No | ||
| 64 | EFNA1 | 8635 | 0.111 | -0.0793 | No | ||
| 65 | HYAL2 | 8662 | 0.108 | -0.0801 | No | ||
| 66 | POU4F3 | 8933 | 0.059 | -0.0943 | No | ||
| 67 | UBTF | 8980 | 0.050 | -0.0965 | No | ||
| 68 | CHD2 | 9028 | 0.042 | -0.0988 | No | ||
| 69 | GRM3 | 9151 | 0.022 | -0.1053 | No | ||
| 70 | PTPRN | 9169 | 0.019 | -0.1061 | No | ||
| 71 | TWIST1 | 9173 | 0.019 | -0.1062 | No | ||
| 72 | SDCCAG8 | 9254 | 0.008 | -0.1105 | No | ||
| 73 | RHOV | 9348 | -0.008 | -0.1155 | No | ||
| 74 | BTBD12 | 9565 | -0.041 | -0.1269 | No | ||
| 75 | UQCRH | 9605 | -0.046 | -0.1288 | No | ||
| 76 | FOSL1 | 10083 | -0.121 | -0.1539 | No | ||
| 77 | CLIC1 | 10110 | -0.126 | -0.1545 | No | ||
| 78 | DDIT3 | 10339 | -0.163 | -0.1659 | No | ||
| 79 | CAMK1D | 10688 | -0.217 | -0.1835 | No | ||
| 80 | STAT5B | 10701 | -0.218 | -0.1828 | No | ||
| 81 | PEX16 | 10979 | -0.257 | -0.1963 | No | ||
| 82 | TFRC | 10982 | -0.257 | -0.1949 | No | ||
| 83 | CNNM1 | 11065 | -0.271 | -0.1977 | No | ||
| 84 | OLFML2A | 11075 | -0.272 | -0.1966 | No | ||
| 85 | DPP3 | 11105 | -0.276 | -0.1965 | No | ||
| 86 | EGR3 | 11317 | -0.308 | -0.2061 | No | ||
| 87 | PLCB2 | 11724 | -0.375 | -0.2259 | No | ||
| 88 | PEX11B | 11756 | -0.380 | -0.2253 | No | ||
| 89 | HOXC4 | 11823 | -0.391 | -0.2265 | No | ||
| 90 | LYL1 | 12059 | -0.430 | -0.2367 | No | ||
| 91 | MDFI | 12244 | -0.459 | -0.2439 | No | ||
| 92 | MAP4K1 | 12297 | -0.468 | -0.2440 | No | ||
| 93 | ADAMTS4 | 12597 | -0.520 | -0.2571 | No | ||
| 94 | LAT | 12618 | -0.523 | -0.2550 | No | ||
| 95 | LYPLA2 | 12660 | -0.529 | -0.2541 | No | ||
| 96 | ARHGDIB | 12754 | -0.547 | -0.2559 | No | ||
| 97 | ATP1A1 | 12805 | -0.555 | -0.2553 | No | ||
| 98 | PIK3CG | 13006 | -0.586 | -0.2626 | No | ||
| 99 | NLK | 13271 | -0.634 | -0.2731 | No | ||
| 100 | IKBKB | 13552 | -0.685 | -0.2842 | No | ||
| 101 | USP5 | 13697 | -0.710 | -0.2878 | No | ||
| 102 | SLC4A2 | 13702 | -0.711 | -0.2838 | No | ||
| 103 | MCTS1 | 13796 | -0.728 | -0.2844 | No | ||
| 104 | TRAPPC3 | 14000 | -0.764 | -0.2909 | No | ||
| 105 | ROM1 | 14166 | -0.798 | -0.2951 | No | ||
| 106 | LANCL1 | 14417 | -0.846 | -0.3036 | Yes | ||
| 107 | ST7L | 14422 | -0.847 | -0.2987 | Yes | ||
| 108 | GGTL3 | 14524 | -0.863 | -0.2990 | Yes | ||
| 109 | NFIA | 14603 | -0.878 | -0.2980 | Yes | ||
| 110 | HMGN2 | 14645 | -0.885 | -0.2950 | Yes | ||
| 111 | PTPN6 | 14685 | -0.894 | -0.2918 | Yes | ||
| 112 | POLD4 | 14732 | -0.904 | -0.2888 | Yes | ||
| 113 | ZHX2 | 14736 | -0.906 | -0.2836 | Yes | ||
| 114 | CAPZA1 | 14759 | -0.909 | -0.2794 | Yes | ||
| 115 | TLN1 | 14857 | -0.932 | -0.2791 | Yes | ||
| 116 | BMP4 | 15124 | -0.993 | -0.2876 | Yes | ||
| 117 | MAPK3 | 15232 | -1.017 | -0.2873 | Yes | ||
| 118 | TPP2 | 15316 | -1.036 | -0.2856 | Yes | ||
| 119 | AGPAT1 | 15386 | -1.053 | -0.2830 | Yes | ||
| 120 | DOK1 | 15402 | -1.057 | -0.2775 | Yes | ||
| 121 | POU6F2 | 15495 | -1.078 | -0.2761 | Yes | ||
| 122 | TGIF2 | 15528 | -1.087 | -0.2713 | Yes | ||
| 123 | RASA1 | 15574 | -1.098 | -0.2672 | Yes | ||
| 124 | SEMA4C | 15588 | -1.101 | -0.2614 | Yes | ||
| 125 | JUNB | 15718 | -1.135 | -0.2616 | Yes | ||
| 126 | C1QTNF6 | 15950 | -1.201 | -0.2669 | Yes | ||
| 127 | TMEM4 | 15966 | -1.204 | -0.2606 | Yes | ||
| 128 | ROBO4 | 15997 | -1.210 | -0.2550 | Yes | ||
| 129 | SRPR | 16003 | -1.212 | -0.2480 | Yes | ||
| 130 | STAT5A | 16004 | -1.212 | -0.2408 | Yes | ||
| 131 | LYN | 16042 | -1.220 | -0.2355 | Yes | ||
| 132 | HCLS1 | 16112 | -1.239 | -0.2318 | Yes | ||
| 133 | GMFB | 16116 | -1.240 | -0.2246 | Yes | ||
| 134 | UBE2L3 | 16319 | -1.300 | -0.2278 | Yes | ||
| 135 | RPS6KA4 | 16321 | -1.301 | -0.2201 | Yes | ||
| 136 | BAZ2A | 16358 | -1.312 | -0.2142 | Yes | ||
| 137 | ITPR3 | 16442 | -1.342 | -0.2107 | Yes | ||
| 138 | FXYD5 | 16472 | -1.350 | -0.2042 | Yes | ||
| 139 | CENTD2 | 16608 | -1.394 | -0.2032 | Yes | ||
| 140 | PIGW | 16652 | -1.411 | -0.1971 | Yes | ||
| 141 | MPL | 16734 | -1.442 | -0.1929 | Yes | ||
| 142 | NTRK3 | 16756 | -1.450 | -0.1853 | Yes | ||
| 143 | NPY | 16977 | -1.532 | -0.1881 | Yes | ||
| 144 | PPP1R9B | 17073 | -1.577 | -0.1839 | Yes | ||
| 145 | SPRED2 | 17144 | -1.603 | -0.1781 | Yes | ||
| 146 | PITX2 | 17203 | -1.627 | -0.1715 | Yes | ||
| 147 | SALL1 | 17215 | -1.632 | -0.1624 | Yes | ||
| 148 | HTF9C | 17223 | -1.635 | -0.1530 | Yes | ||
| 149 | CREB3 | 17287 | -1.658 | -0.1465 | Yes | ||
| 150 | RASAL1 | 17309 | -1.669 | -0.1377 | Yes | ||
| 151 | ERG | 17385 | -1.700 | -0.1316 | Yes | ||
| 152 | DEF6 | 17448 | -1.731 | -0.1246 | Yes | ||
| 153 | RANBP1 | 17466 | -1.738 | -0.1152 | Yes | ||
| 154 | FBS1 | 17496 | -1.753 | -0.1063 | Yes | ||
| 155 | CORO1A | 17617 | -1.818 | -0.1019 | Yes | ||
| 156 | EBAG9 | 17622 | -1.821 | -0.0913 | Yes | ||
| 157 | SNCG | 17632 | -1.827 | -0.0809 | Yes | ||
| 158 | HMGA1 | 17771 | -1.910 | -0.0769 | Yes | ||
| 159 | RBBP7 | 17815 | -1.941 | -0.0677 | Yes | ||
| 160 | HIC1 | 17921 | -2.010 | -0.0614 | Yes | ||
| 161 | FGR | 18013 | -2.084 | -0.0539 | Yes | ||
| 162 | MID1 | 18092 | -2.165 | -0.0452 | Yes | ||
| 163 | SH2D2A | 18208 | -2.294 | -0.0377 | Yes | ||
| 164 | FBXO36 | 18219 | -2.311 | -0.0244 | Yes | ||
| 165 | ZFPM1 | 18225 | -2.318 | -0.0109 | Yes | ||
| 166 | FLT1 | 18379 | -2.543 | -0.0040 | Yes | ||
| 167 | RGS3 | 18468 | -2.810 | 0.0080 | Yes |
