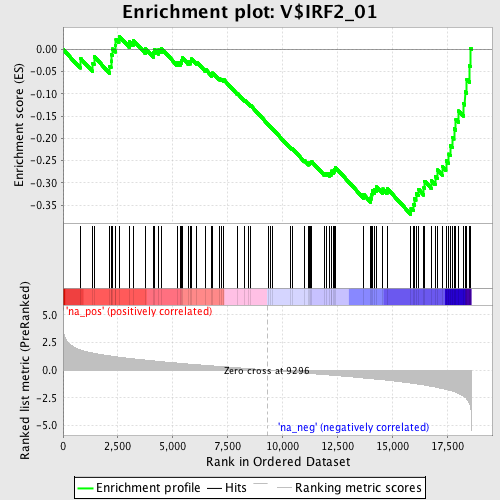
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$IRF2_01 |
| Enrichment Score (ES) | -0.3711832 |
| Normalized Enrichment Score (NES) | -1.6127163 |
| Nominal p-value | 0.0017953322 |
| FDR q-value | 0.66228414 |
| FWER p-Value | 0.788 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VGLL4 | 777 | 1.832 | -0.0193 | No | ||
| 2 | PSMA3 | 1354 | 1.545 | -0.0313 | No | ||
| 3 | MAPK6 | 1414 | 1.518 | -0.0158 | No | ||
| 4 | BNC2 | 2122 | 1.280 | -0.0381 | No | ||
| 5 | SLC38A2 | 2186 | 1.262 | -0.0259 | No | ||
| 6 | ATF3 | 2208 | 1.257 | -0.0116 | No | ||
| 7 | E2F3 | 2258 | 1.247 | 0.0012 | No | ||
| 8 | DAP3 | 2392 | 1.209 | 0.0089 | No | ||
| 9 | CDK6 | 2407 | 1.205 | 0.0230 | No | ||
| 10 | LDB2 | 2548 | 1.170 | 0.0299 | No | ||
| 11 | USP18 | 3021 | 1.055 | 0.0175 | No | ||
| 12 | NR3C2 | 3191 | 1.019 | 0.0209 | No | ||
| 13 | PIGR | 3747 | 0.906 | 0.0022 | No | ||
| 14 | KCNE4 | 4116 | 0.831 | -0.0074 | No | ||
| 15 | HOXA6 | 4170 | 0.818 | -0.0002 | No | ||
| 16 | THRAP1 | 4365 | 0.784 | -0.0010 | No | ||
| 17 | RIMS2 | 4486 | 0.761 | 0.0019 | No | ||
| 18 | ASH1L | 5207 | 0.632 | -0.0291 | No | ||
| 19 | IFNB1 | 5334 | 0.610 | -0.0284 | No | ||
| 20 | ARHGAP5 | 5409 | 0.595 | -0.0250 | No | ||
| 21 | NDN | 5428 | 0.592 | -0.0187 | No | ||
| 22 | PGM5 | 5715 | 0.549 | -0.0273 | No | ||
| 23 | ESR1 | 5818 | 0.528 | -0.0263 | No | ||
| 24 | LIF | 5844 | 0.525 | -0.0212 | No | ||
| 25 | PRDM16 | 6095 | 0.486 | -0.0287 | No | ||
| 26 | BHLHB5 | 6484 | 0.431 | -0.0443 | No | ||
| 27 | DNASE1L3 | 6784 | 0.385 | -0.0557 | No | ||
| 28 | PLXNC1 | 6818 | 0.381 | -0.0528 | No | ||
| 29 | PDGFC | 7118 | 0.337 | -0.0647 | No | ||
| 30 | IL27 | 7230 | 0.323 | -0.0667 | No | ||
| 31 | MCM8 | 7327 | 0.310 | -0.0681 | No | ||
| 32 | CUL2 | 7954 | 0.211 | -0.0993 | No | ||
| 33 | PRDX5 | 8272 | 0.163 | -0.1144 | No | ||
| 34 | ASPA | 8427 | 0.142 | -0.1209 | No | ||
| 35 | DLX4 | 8561 | 0.122 | -0.1266 | No | ||
| 36 | MITF | 9345 | -0.008 | -0.1687 | No | ||
| 37 | PROX1 | 9466 | -0.026 | -0.1749 | No | ||
| 38 | CXCL10 | 9539 | -0.037 | -0.1783 | No | ||
| 39 | NT5C3 | 10359 | -0.167 | -0.2205 | No | ||
| 40 | NPR3 | 10436 | -0.178 | -0.2224 | No | ||
| 41 | MAP3K11 | 11008 | -0.261 | -0.2499 | No | ||
| 42 | BBX | 11197 | -0.289 | -0.2565 | No | ||
| 43 | RFX4 | 11238 | -0.296 | -0.2550 | No | ||
| 44 | DDX58 | 11276 | -0.301 | -0.2533 | No | ||
| 45 | USF1 | 11336 | -0.311 | -0.2527 | No | ||
| 46 | UBD | 11928 | -0.409 | -0.2795 | No | ||
| 47 | CSAD | 12009 | -0.422 | -0.2786 | No | ||
| 48 | EIF4A2 | 12130 | -0.442 | -0.2796 | No | ||
| 49 | PSMB8 | 12213 | -0.454 | -0.2785 | No | ||
| 50 | IL11 | 12223 | -0.455 | -0.2733 | No | ||
| 51 | BLK | 12329 | -0.475 | -0.2731 | No | ||
| 52 | TGFB3 | 12380 | -0.483 | -0.2699 | No | ||
| 53 | BST2 | 12400 | -0.486 | -0.2649 | No | ||
| 54 | USP5 | 13697 | -0.710 | -0.3261 | No | ||
| 55 | TBX1 | 14022 | -0.769 | -0.3341 | No | ||
| 56 | SIPA1 | 14069 | -0.779 | -0.3269 | No | ||
| 57 | MSX1 | 14079 | -0.782 | -0.3178 | No | ||
| 58 | DLX1 | 14195 | -0.802 | -0.3141 | No | ||
| 59 | BCL11A | 14288 | -0.819 | -0.3089 | No | ||
| 60 | TMPRSS5 | 14577 | -0.874 | -0.3137 | No | ||
| 61 | TAP1 | 14766 | -0.910 | -0.3126 | No | ||
| 62 | GLRA1 | 15853 | -1.172 | -0.3567 | Yes | ||
| 63 | HPCAL1 | 15967 | -1.204 | -0.3479 | Yes | ||
| 64 | ARHGEF6 | 16015 | -1.215 | -0.3355 | Yes | ||
| 65 | DLG1 | 16084 | -1.232 | -0.3239 | Yes | ||
| 66 | GSDMDC1 | 16189 | -1.262 | -0.3140 | Yes | ||
| 67 | KARS | 16443 | -1.342 | -0.3111 | Yes | ||
| 68 | FXYD5 | 16472 | -1.350 | -0.2959 | Yes | ||
| 69 | UPF2 | 16798 | -1.467 | -0.2954 | Yes | ||
| 70 | TERF2IP | 16959 | -1.526 | -0.2852 | Yes | ||
| 71 | BAT5 | 17053 | -1.566 | -0.2708 | Yes | ||
| 72 | ROCK1 | 17278 | -1.656 | -0.2625 | Yes | ||
| 73 | DUSP10 | 17452 | -1.732 | -0.2504 | Yes | ||
| 74 | TCIRG1 | 17580 | -1.796 | -0.2351 | Yes | ||
| 75 | OTX1 | 17648 | -1.837 | -0.2161 | Yes | ||
| 76 | EGFL6 | 17744 | -1.894 | -0.1978 | Yes | ||
| 77 | REL | 17816 | -1.941 | -0.1777 | Yes | ||
| 78 | TAPBP | 17901 | -1.994 | -0.1576 | Yes | ||
| 79 | TNFSF13B | 18023 | -2.096 | -0.1382 | Yes | ||
| 80 | IL15RA | 18243 | -2.337 | -0.1212 | Yes | ||
| 81 | PSMB9 | 18317 | -2.445 | -0.0950 | Yes | ||
| 82 | PSME1 | 18400 | -2.603 | -0.0673 | Yes | ||
| 83 | TRPM3 | 18525 | -3.023 | -0.0366 | Yes | ||
| 84 | B2M | 18574 | -3.363 | 0.0023 | Yes |
