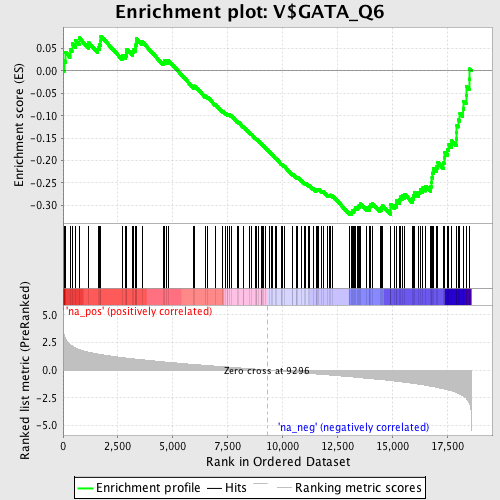
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$GATA_Q6 |
| Enrichment Score (ES) | -0.3203746 |
| Normalized Enrichment Score (NES) | -1.5068 |
| Nominal p-value | 0.0088495575 |
| FDR q-value | 0.46877035 |
| FWER p-Value | 0.988 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SYT7 | 44 | 3.220 | 0.0229 | No | ||
| 2 | LMO2 | 109 | 2.847 | 0.0417 | No | ||
| 3 | KLF12 | 314 | 2.309 | 0.0488 | No | ||
| 4 | H1F0 | 409 | 2.177 | 0.0608 | No | ||
| 5 | FOSL2 | 561 | 2.012 | 0.0684 | No | ||
| 6 | SFRP5 | 731 | 1.861 | 0.0738 | No | ||
| 7 | RGN | 1163 | 1.623 | 0.0632 | No | ||
| 8 | PCDH9 | 1590 | 1.446 | 0.0515 | No | ||
| 9 | JPH1 | 1655 | 1.422 | 0.0592 | No | ||
| 10 | CLU | 1718 | 1.402 | 0.0668 | No | ||
| 11 | PDGFRA | 1722 | 1.401 | 0.0777 | No | ||
| 12 | RAP1B | 2690 | 1.135 | 0.0342 | No | ||
| 13 | TRPS1 | 2829 | 1.097 | 0.0354 | No | ||
| 14 | MAP4K3 | 2891 | 1.081 | 0.0405 | No | ||
| 15 | KIRREL2 | 2903 | 1.079 | 0.0484 | No | ||
| 16 | SLITRK2 | 3161 | 1.026 | 0.0425 | No | ||
| 17 | TBX5 | 3198 | 1.017 | 0.0486 | No | ||
| 18 | ROBO1 | 3305 | 0.995 | 0.0506 | No | ||
| 19 | PKLR | 3313 | 0.994 | 0.0581 | No | ||
| 20 | JUN | 3331 | 0.989 | 0.0649 | No | ||
| 21 | ICAM4 | 3346 | 0.985 | 0.0719 | No | ||
| 22 | ADM | 3597 | 0.938 | 0.0657 | No | ||
| 23 | MASP1 | 4563 | 0.744 | 0.0193 | No | ||
| 24 | NRXN3 | 4602 | 0.737 | 0.0230 | No | ||
| 25 | GATA1 | 4707 | 0.719 | 0.0230 | No | ||
| 26 | ESRRG | 4810 | 0.702 | 0.0230 | No | ||
| 27 | ISL1 | 5949 | 0.509 | -0.0346 | No | ||
| 28 | GPR44 | 6004 | 0.500 | -0.0336 | No | ||
| 29 | MS4A2 | 6476 | 0.432 | -0.0557 | No | ||
| 30 | ZNF219 | 6596 | 0.412 | -0.0589 | No | ||
| 31 | LHX6 | 6933 | 0.362 | -0.0743 | No | ||
| 32 | SPRY4 | 7278 | 0.318 | -0.0904 | No | ||
| 33 | MOGAT2 | 7392 | 0.302 | -0.0941 | No | ||
| 34 | CXCL13 | 7505 | 0.285 | -0.0980 | No | ||
| 35 | CS | 7514 | 0.283 | -0.0962 | No | ||
| 36 | ABCA12 | 7568 | 0.275 | -0.0969 | No | ||
| 37 | KCNJ15 | 7670 | 0.260 | -0.1003 | No | ||
| 38 | GFRA3 | 7943 | 0.213 | -0.1134 | No | ||
| 39 | CPA1 | 8001 | 0.205 | -0.1149 | No | ||
| 40 | LCAT | 8232 | 0.170 | -0.1260 | No | ||
| 41 | ADORA3 | 8237 | 0.169 | -0.1249 | No | ||
| 42 | RHD | 8494 | 0.131 | -0.1377 | No | ||
| 43 | PPARGC1A | 8583 | 0.121 | -0.1415 | No | ||
| 44 | RBPMS | 8750 | 0.092 | -0.1498 | No | ||
| 45 | IL7 | 8816 | 0.081 | -0.1526 | No | ||
| 46 | PLAC1 | 8829 | 0.079 | -0.1527 | No | ||
| 47 | GIP | 8902 | 0.065 | -0.1561 | No | ||
| 48 | TSHB | 9027 | 0.042 | -0.1624 | No | ||
| 49 | IL22 | 9033 | 0.040 | -0.1624 | No | ||
| 50 | PFKFB1 | 9042 | 0.038 | -0.1625 | No | ||
| 51 | PNLIPRP2 | 9098 | 0.029 | -0.1653 | No | ||
| 52 | SLC4A1 | 9112 | 0.028 | -0.1658 | No | ||
| 53 | IGFBP5 | 9208 | 0.014 | -0.1708 | No | ||
| 54 | GP2 | 9397 | -0.017 | -0.1808 | No | ||
| 55 | CDC42EP3 | 9517 | -0.033 | -0.1870 | No | ||
| 56 | FLI1 | 9557 | -0.040 | -0.1888 | No | ||
| 57 | FMO1 | 9698 | -0.060 | -0.1959 | No | ||
| 58 | HOXD1 | 9708 | -0.061 | -0.1959 | No | ||
| 59 | CCRN4L | 9966 | -0.101 | -0.2091 | No | ||
| 60 | GATA6 | 9975 | -0.102 | -0.2087 | No | ||
| 61 | BMP6 | 10001 | -0.106 | -0.2092 | No | ||
| 62 | AMFR | 10085 | -0.121 | -0.2127 | No | ||
| 63 | HAMP | 10459 | -0.182 | -0.2315 | No | ||
| 64 | SIX1 | 10474 | -0.184 | -0.2308 | No | ||
| 65 | CNN1 | 10644 | -0.209 | -0.2383 | No | ||
| 66 | MOS | 10655 | -0.211 | -0.2372 | No | ||
| 67 | PLXNB3 | 10680 | -0.215 | -0.2368 | No | ||
| 68 | CD34 | 10850 | -0.239 | -0.2441 | No | ||
| 69 | CREB5 | 11005 | -0.260 | -0.2504 | No | ||
| 70 | FHL2 | 11063 | -0.270 | -0.2514 | No | ||
| 71 | MSX2 | 11194 | -0.289 | -0.2561 | No | ||
| 72 | FTH1 | 11213 | -0.292 | -0.2548 | No | ||
| 73 | TRIM10 | 11389 | -0.320 | -0.2618 | No | ||
| 74 | EN1 | 11527 | -0.344 | -0.2665 | No | ||
| 75 | EPX | 11537 | -0.346 | -0.2643 | No | ||
| 76 | VPS18 | 11583 | -0.353 | -0.2639 | No | ||
| 77 | BPGM | 11657 | -0.364 | -0.2650 | No | ||
| 78 | PHOX2A | 11788 | -0.385 | -0.2690 | No | ||
| 79 | ENO2 | 11851 | -0.396 | -0.2693 | No | ||
| 80 | LYL1 | 12059 | -0.430 | -0.2771 | No | ||
| 81 | BSN | 12134 | -0.443 | -0.2777 | No | ||
| 82 | USH1G | 12185 | -0.449 | -0.2768 | No | ||
| 83 | PLAGL2 | 12290 | -0.467 | -0.2788 | No | ||
| 84 | SLC18A1 | 13059 | -0.594 | -0.3157 | Yes | ||
| 85 | KCNK3 | 13146 | -0.608 | -0.3156 | Yes | ||
| 86 | CACNA2D3 | 13169 | -0.614 | -0.3120 | Yes | ||
| 87 | FOXP2 | 13256 | -0.631 | -0.3117 | Yes | ||
| 88 | HOXA1 | 13298 | -0.639 | -0.3089 | Yes | ||
| 89 | EDN1 | 13319 | -0.641 | -0.3049 | Yes | ||
| 90 | INHA | 13406 | -0.657 | -0.3044 | Yes | ||
| 91 | OTOP2 | 13446 | -0.663 | -0.3014 | Yes | ||
| 92 | FABP2 | 13507 | -0.675 | -0.2993 | Yes | ||
| 93 | NR5A2 | 13558 | -0.686 | -0.2966 | Yes | ||
| 94 | EPOR | 13827 | -0.732 | -0.3054 | Yes | ||
| 95 | DSPP | 13948 | -0.754 | -0.3060 | Yes | ||
| 96 | B3GALT2 | 13981 | -0.760 | -0.3018 | Yes | ||
| 97 | FHL3 | 14018 | -0.768 | -0.2977 | Yes | ||
| 98 | BIN1 | 14108 | -0.786 | -0.2963 | Yes | ||
| 99 | GUCA2A | 14445 | -0.850 | -0.3079 | Yes | ||
| 100 | MYCT1 | 14508 | -0.860 | -0.3045 | Yes | ||
| 101 | TNXB | 14570 | -0.872 | -0.3009 | Yes | ||
| 102 | CAPN1 | 14921 | -0.946 | -0.3125 | Yes | ||
| 103 | TIAL1 | 14923 | -0.948 | -0.3051 | Yes | ||
| 104 | WNT11 | 14924 | -0.948 | -0.2976 | Yes | ||
| 105 | USP15 | 15110 | -0.989 | -0.2999 | Yes | ||
| 106 | CCL27 | 15187 | -1.007 | -0.2961 | Yes | ||
| 107 | TRIM15 | 15207 | -1.012 | -0.2892 | Yes | ||
| 108 | AQP2 | 15331 | -1.040 | -0.2877 | Yes | ||
| 109 | BMP10 | 15363 | -1.047 | -0.2812 | Yes | ||
| 110 | HCN1 | 15458 | -1.068 | -0.2779 | Yes | ||
| 111 | POFUT1 | 15571 | -1.098 | -0.2753 | Yes | ||
| 112 | PVRL2 | 15921 | -1.193 | -0.2849 | Yes | ||
| 113 | SLC26A9 | 15973 | -1.205 | -0.2782 | Yes | ||
| 114 | KRT20 | 16022 | -1.217 | -0.2712 | Yes | ||
| 115 | PLA2G1B | 16202 | -1.265 | -0.2710 | Yes | ||
| 116 | ELAVL4 | 16268 | -1.285 | -0.2644 | Yes | ||
| 117 | NRP2 | 16398 | -1.325 | -0.2610 | Yes | ||
| 118 | DCX | 16529 | -1.370 | -0.2573 | Yes | ||
| 119 | PIP5K1B | 16753 | -1.450 | -0.2580 | Yes | ||
| 120 | COL4A3 | 16780 | -1.460 | -0.2480 | Yes | ||
| 121 | WDR23 | 16811 | -1.471 | -0.2381 | Yes | ||
| 122 | PAX2 | 16835 | -1.479 | -0.2277 | Yes | ||
| 123 | COL4A4 | 16868 | -1.494 | -0.2177 | Yes | ||
| 124 | HMBS | 17012 | -1.549 | -0.2133 | Yes | ||
| 125 | PPT2 | 17081 | -1.582 | -0.2046 | Yes | ||
| 126 | SUV39H1 | 17331 | -1.679 | -0.2049 | Yes | ||
| 127 | PLAG1 | 17368 | -1.695 | -0.1936 | Yes | ||
| 128 | ERG | 17385 | -1.700 | -0.1811 | Yes | ||
| 129 | PVRL4 | 17535 | -1.768 | -0.1753 | Yes | ||
| 130 | GATM | 17578 | -1.796 | -0.1635 | Yes | ||
| 131 | SPINK4 | 17712 | -1.873 | -0.1560 | Yes | ||
| 132 | HIC1 | 17921 | -2.010 | -0.1515 | Yes | ||
| 133 | HOXB6 | 17935 | -2.018 | -0.1364 | Yes | ||
| 134 | SLC26A7 | 17945 | -2.022 | -0.1210 | Yes | ||
| 135 | TNFSF13B | 18023 | -2.096 | -0.1087 | Yes | ||
| 136 | PNLIPRP1 | 18085 | -2.159 | -0.0951 | Yes | ||
| 137 | ZFPM1 | 18225 | -2.318 | -0.0844 | Yes | ||
| 138 | XPO6 | 18257 | -2.357 | -0.0676 | Yes | ||
| 139 | AMOT | 18369 | -2.527 | -0.0538 | Yes | ||
| 140 | CPOX | 18386 | -2.563 | -0.0346 | Yes | ||
| 141 | CHCHD7 | 18504 | -2.927 | -0.0180 | Yes | ||
| 142 | POU4F2 | 18534 | -3.059 | 0.0044 | Yes |
