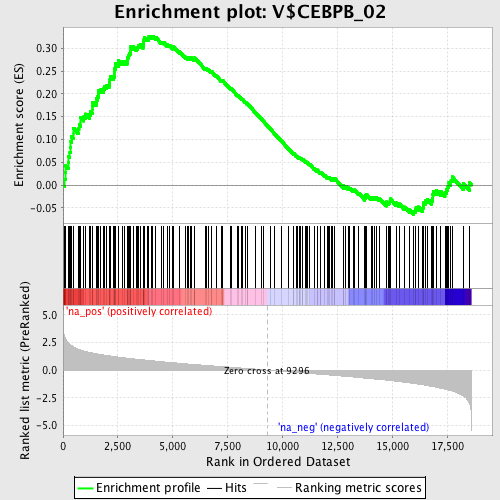
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CEBPB_02 |
| Enrichment Score (ES) | 0.3265457 |
| Normalized Enrichment Score (NES) | 1.659834 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.49485052 |
| FWER p-Value | 0.538 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UBQLN1 | 75 | 3.000 | 0.0127 | Yes | ||
| 2 | PCDH7 | 101 | 2.871 | 0.0274 | Yes | ||
| 3 | DUSP1 | 112 | 2.834 | 0.0427 | Yes | ||
| 4 | PER1 | 231 | 2.454 | 0.0500 | Yes | ||
| 5 | VNN3 | 245 | 2.413 | 0.0627 | Yes | ||
| 6 | FOXP1 | 309 | 2.323 | 0.0723 | Yes | ||
| 7 | SLC6A4 | 343 | 2.268 | 0.0832 | Yes | ||
| 8 | NEK6 | 356 | 2.236 | 0.0950 | Yes | ||
| 9 | FGA | 379 | 2.213 | 0.1062 | Yes | ||
| 10 | CCND2 | 472 | 2.098 | 0.1129 | Yes | ||
| 11 | TOP1 | 474 | 2.093 | 0.1246 | Yes | ||
| 12 | H3F3B | 695 | 1.889 | 0.1232 | Yes | ||
| 13 | FIGN | 725 | 1.869 | 0.1320 | Yes | ||
| 14 | SYNCRIP | 800 | 1.816 | 0.1382 | Yes | ||
| 15 | SOCS2 | 801 | 1.816 | 0.1483 | Yes | ||
| 16 | LUZP1 | 949 | 1.726 | 0.1500 | Yes | ||
| 17 | DMD | 1017 | 1.693 | 0.1558 | Yes | ||
| 18 | ONECUT2 | 1211 | 1.608 | 0.1543 | Yes | ||
| 19 | CSNK1E | 1242 | 1.594 | 0.1616 | Yes | ||
| 20 | PPARG | 1339 | 1.551 | 0.1650 | Yes | ||
| 21 | MTF1 | 1343 | 1.548 | 0.1735 | Yes | ||
| 22 | CACNB2 | 1351 | 1.546 | 0.1818 | Yes | ||
| 23 | HNRPR | 1504 | 1.479 | 0.1818 | Yes | ||
| 24 | SLIT3 | 1512 | 1.478 | 0.1897 | Yes | ||
| 25 | KPNA3 | 1583 | 1.448 | 0.1940 | Yes | ||
| 26 | SLC12A1 | 1610 | 1.438 | 0.2006 | Yes | ||
| 27 | NFKBIA | 1624 | 1.433 | 0.2079 | Yes | ||
| 28 | PDGFRA | 1722 | 1.401 | 0.2105 | Yes | ||
| 29 | EFNA5 | 1856 | 1.361 | 0.2108 | Yes | ||
| 30 | EIF4A1 | 1897 | 1.349 | 0.2162 | Yes | ||
| 31 | ACLY | 1982 | 1.320 | 0.2190 | Yes | ||
| 32 | RRBP1 | 2094 | 1.288 | 0.2202 | Yes | ||
| 33 | BNC2 | 2122 | 1.280 | 0.2259 | Yes | ||
| 34 | G0S2 | 2134 | 1.275 | 0.2324 | Yes | ||
| 35 | ARF6 | 2158 | 1.269 | 0.2382 | Yes | ||
| 36 | ATAD2 | 2289 | 1.238 | 0.2381 | Yes | ||
| 37 | RHOB | 2329 | 1.229 | 0.2429 | Yes | ||
| 38 | THRA | 2334 | 1.228 | 0.2495 | Yes | ||
| 39 | RUVBL2 | 2347 | 1.224 | 0.2557 | Yes | ||
| 40 | TBR1 | 2390 | 1.210 | 0.2602 | Yes | ||
| 41 | CITED1 | 2402 | 1.206 | 0.2663 | Yes | ||
| 42 | ARNT | 2517 | 1.176 | 0.2667 | Yes | ||
| 43 | UBE2E2 | 2530 | 1.174 | 0.2726 | Yes | ||
| 44 | CCL3 | 2684 | 1.138 | 0.2707 | Yes | ||
| 45 | SLC7A11 | 2790 | 1.105 | 0.2711 | Yes | ||
| 46 | MPP6 | 2913 | 1.075 | 0.2705 | Yes | ||
| 47 | PFN2 | 2935 | 1.071 | 0.2754 | Yes | ||
| 48 | SERTAD4 | 2951 | 1.067 | 0.2805 | Yes | ||
| 49 | TCF12 | 2991 | 1.060 | 0.2843 | Yes | ||
| 50 | PHF16 | 3013 | 1.057 | 0.2891 | Yes | ||
| 51 | MAP2K6 | 3057 | 1.047 | 0.2926 | Yes | ||
| 52 | PDAP1 | 3081 | 1.041 | 0.2972 | Yes | ||
| 53 | ASCL2 | 3082 | 1.041 | 0.3030 | Yes | ||
| 54 | FMO2 | 3186 | 1.020 | 0.3031 | Yes | ||
| 55 | FST | 3339 | 0.988 | 0.3004 | Yes | ||
| 56 | MAP4K4 | 3405 | 0.972 | 0.3023 | Yes | ||
| 57 | UGP2 | 3426 | 0.969 | 0.3066 | Yes | ||
| 58 | SPRED1 | 3505 | 0.955 | 0.3077 | Yes | ||
| 59 | LEP | 3643 | 0.929 | 0.3055 | Yes | ||
| 60 | KCNE3 | 3662 | 0.924 | 0.3096 | Yes | ||
| 61 | TOB1 | 3667 | 0.923 | 0.3146 | Yes | ||
| 62 | MBNL1 | 3684 | 0.919 | 0.3188 | Yes | ||
| 63 | AQP9 | 3701 | 0.915 | 0.3231 | Yes | ||
| 64 | RPS21 | 3834 | 0.887 | 0.3209 | Yes | ||
| 65 | RPA3 | 3907 | 0.872 | 0.3218 | Yes | ||
| 66 | DYRK3 | 3911 | 0.870 | 0.3265 | Yes | ||
| 67 | PTPN12 | 4017 | 0.850 | 0.3256 | No | ||
| 68 | CDKN1B | 4088 | 0.836 | 0.3265 | No | ||
| 69 | PCTP | 4214 | 0.810 | 0.3242 | No | ||
| 70 | TRIB1 | 4490 | 0.760 | 0.3135 | No | ||
| 71 | SMAD6 | 4570 | 0.743 | 0.3134 | No | ||
| 72 | BCL6 | 4734 | 0.715 | 0.3086 | No | ||
| 73 | S100A9 | 4836 | 0.698 | 0.3070 | No | ||
| 74 | SMARCA1 | 5000 | 0.667 | 0.3019 | No | ||
| 75 | PPM1B | 5041 | 0.662 | 0.3034 | No | ||
| 76 | HAVCR1 | 5310 | 0.614 | 0.2923 | No | ||
| 77 | H2AFZ | 5580 | 0.570 | 0.2809 | No | ||
| 78 | PDGFB | 5688 | 0.553 | 0.2781 | No | ||
| 79 | PPL | 5710 | 0.550 | 0.2801 | No | ||
| 80 | ESR1 | 5818 | 0.528 | 0.2772 | No | ||
| 81 | STX18 | 5841 | 0.526 | 0.2790 | No | ||
| 82 | ITGA11 | 5966 | 0.506 | 0.2751 | No | ||
| 83 | TMPRSS4 | 5992 | 0.502 | 0.2765 | No | ||
| 84 | PPP1CB | 6002 | 0.500 | 0.2788 | No | ||
| 85 | CSMD3 | 6478 | 0.432 | 0.2555 | No | ||
| 86 | ASAH2 | 6533 | 0.423 | 0.2549 | No | ||
| 87 | CP | 6615 | 0.409 | 0.2528 | No | ||
| 88 | HIST1H1C | 6747 | 0.391 | 0.2479 | No | ||
| 89 | PRDX3 | 6755 | 0.389 | 0.2497 | No | ||
| 90 | P2RY4 | 6983 | 0.355 | 0.2393 | No | ||
| 91 | IL27 | 7230 | 0.323 | 0.2278 | No | ||
| 92 | CYP24A1 | 7244 | 0.322 | 0.2289 | No | ||
| 93 | SPRY4 | 7278 | 0.318 | 0.2289 | No | ||
| 94 | PDGFRL | 7612 | 0.269 | 0.2123 | No | ||
| 95 | FCER1G | 7652 | 0.263 | 0.2116 | No | ||
| 96 | SDPR | 7957 | 0.210 | 0.1963 | No | ||
| 97 | LMO4 | 7980 | 0.207 | 0.1963 | No | ||
| 98 | KCNH7 | 8109 | 0.187 | 0.1904 | No | ||
| 99 | SERPINF1 | 8164 | 0.179 | 0.1885 | No | ||
| 100 | PIP5K1A | 8304 | 0.158 | 0.1818 | No | ||
| 101 | MAP3K3 | 8387 | 0.147 | 0.1782 | No | ||
| 102 | RGR | 8395 | 0.145 | 0.1786 | No | ||
| 103 | RBPMS | 8750 | 0.092 | 0.1599 | No | ||
| 104 | CHD2 | 9028 | 0.042 | 0.1451 | No | ||
| 105 | BDKRB1 | 9148 | 0.022 | 0.1388 | No | ||
| 106 | SLC19A3 | 9433 | -0.021 | 0.1235 | No | ||
| 107 | BNIP3L | 9651 | -0.053 | 0.1120 | No | ||
| 108 | ELAVL2 | 9967 | -0.101 | 0.0955 | No | ||
| 109 | NHLH1 | 10277 | -0.151 | 0.0796 | No | ||
| 110 | PDCL | 10514 | -0.189 | 0.0678 | No | ||
| 111 | BMF | 10516 | -0.189 | 0.0689 | No | ||
| 112 | IL1RAPL2 | 10633 | -0.207 | 0.0637 | No | ||
| 113 | SP8 | 10691 | -0.217 | 0.0618 | No | ||
| 114 | PCF11 | 10753 | -0.227 | 0.0598 | No | ||
| 115 | LEMD2 | 10787 | -0.232 | 0.0593 | No | ||
| 116 | MRC2 | 10826 | -0.237 | 0.0586 | No | ||
| 117 | RG9MTD2 | 10897 | -0.244 | 0.0561 | No | ||
| 118 | RAD23B | 10914 | -0.247 | 0.0566 | No | ||
| 119 | PHOX2B | 11066 | -0.271 | 0.0500 | No | ||
| 120 | C1QL1 | 11082 | -0.273 | 0.0507 | No | ||
| 121 | SEPHS2 | 11116 | -0.278 | 0.0504 | No | ||
| 122 | RFX4 | 11238 | -0.296 | 0.0455 | No | ||
| 123 | EHF | 11440 | -0.330 | 0.0365 | No | ||
| 124 | ZADH2 | 11575 | -0.353 | 0.0312 | No | ||
| 125 | KCNJ2 | 11576 | -0.353 | 0.0331 | No | ||
| 126 | MEOX2 | 11739 | -0.377 | 0.0265 | No | ||
| 127 | ERF | 11745 | -0.378 | 0.0283 | No | ||
| 128 | PAFAH2 | 11920 | -0.408 | 0.0211 | No | ||
| 129 | ALDH1A2 | 12033 | -0.426 | 0.0174 | No | ||
| 130 | STAT3 | 12107 | -0.438 | 0.0159 | No | ||
| 131 | EIF4A2 | 12130 | -0.442 | 0.0172 | No | ||
| 132 | ADRB2 | 12252 | -0.460 | 0.0132 | No | ||
| 133 | HOXA5 | 12271 | -0.463 | 0.0148 | No | ||
| 134 | MYO1C | 12373 | -0.482 | 0.0120 | No | ||
| 135 | TGFB3 | 12380 | -0.483 | 0.0144 | No | ||
| 136 | CASK | 12781 | -0.552 | -0.0042 | No | ||
| 137 | SERPINA7 | 12785 | -0.552 | -0.0013 | No | ||
| 138 | PLA2G4A | 12892 | -0.569 | -0.0039 | No | ||
| 139 | SMAD1 | 12994 | -0.584 | -0.0061 | No | ||
| 140 | RIN3 | 13073 | -0.596 | -0.0070 | No | ||
| 141 | SULF1 | 13220 | -0.623 | -0.0114 | No | ||
| 142 | PRG4 | 13273 | -0.634 | -0.0107 | No | ||
| 143 | CHRM1 | 13478 | -0.669 | -0.0180 | No | ||
| 144 | PDE3B | 13758 | -0.721 | -0.0291 | No | ||
| 145 | HOXC13 | 13772 | -0.724 | -0.0258 | No | ||
| 146 | FGB | 13778 | -0.725 | -0.0220 | No | ||
| 147 | CUEDC1 | 13823 | -0.732 | -0.0203 | No | ||
| 148 | PDE4D | 14038 | -0.772 | -0.0276 | No | ||
| 149 | GYS1 | 14112 | -0.787 | -0.0272 | No | ||
| 150 | DLX1 | 14195 | -0.802 | -0.0271 | No | ||
| 151 | BCL11A | 14288 | -0.819 | -0.0276 | No | ||
| 152 | NFIL3 | 14425 | -0.847 | -0.0302 | No | ||
| 153 | FGF14 | 14719 | -0.900 | -0.0411 | No | ||
| 154 | SOX5 | 14737 | -0.906 | -0.0369 | No | ||
| 155 | MOSPD2 | 14846 | -0.930 | -0.0376 | No | ||
| 156 | NFE2L2 | 14896 | -0.939 | -0.0350 | No | ||
| 157 | SMARCAD1 | 14908 | -0.942 | -0.0303 | No | ||
| 158 | EDN2 | 15174 | -1.004 | -0.0391 | No | ||
| 159 | GLP2R | 15323 | -1.038 | -0.0413 | No | ||
| 160 | ARRDC3 | 15561 | -1.096 | -0.0481 | No | ||
| 161 | CDKL5 | 15796 | -1.156 | -0.0543 | No | ||
| 162 | CBX4 | 15986 | -1.208 | -0.0578 | No | ||
| 163 | NDRG1 | 16045 | -1.221 | -0.0541 | No | ||
| 164 | DBH | 16082 | -1.232 | -0.0492 | No | ||
| 165 | WNT5A | 16191 | -1.262 | -0.0480 | No | ||
| 166 | ANGPT1 | 16386 | -1.321 | -0.0512 | No | ||
| 167 | GOT1 | 16409 | -1.327 | -0.0450 | No | ||
| 168 | ITPR3 | 16442 | -1.342 | -0.0392 | No | ||
| 169 | ASGR1 | 16500 | -1.360 | -0.0347 | No | ||
| 170 | IL19 | 16585 | -1.387 | -0.0315 | No | ||
| 171 | COL4A3 | 16780 | -1.460 | -0.0339 | No | ||
| 172 | SLC25A12 | 16838 | -1.480 | -0.0287 | No | ||
| 173 | PLA2G2E | 16853 | -1.486 | -0.0211 | No | ||
| 174 | COL4A4 | 16868 | -1.494 | -0.0136 | No | ||
| 175 | TRPM1 | 17009 | -1.548 | -0.0125 | No | ||
| 176 | PITX2 | 17203 | -1.627 | -0.0139 | No | ||
| 177 | SOX10 | 17406 | -1.709 | -0.0153 | No | ||
| 178 | PKNOX2 | 17469 | -1.739 | -0.0089 | No | ||
| 179 | FBXW7 | 17539 | -1.772 | -0.0028 | No | ||
| 180 | TAC1 | 17583 | -1.798 | 0.0049 | No | ||
| 181 | BDNF | 17678 | -1.854 | 0.0102 | No | ||
| 182 | DDR2 | 17731 | -1.886 | 0.0179 | No | ||
| 183 | PRLR | 18250 | -2.344 | 0.0029 | No | ||
| 184 | RAB3IP | 18527 | -3.029 | 0.0048 | No |
