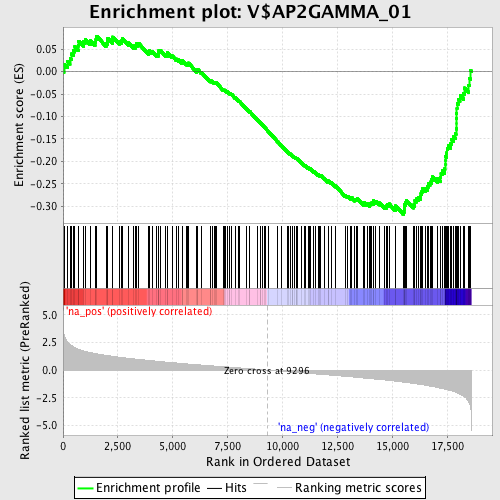
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AP2GAMMA_01 |
| Enrichment Score (ES) | -0.31803873 |
| Normalized Enrichment Score (NES) | -1.5398118 |
| Nominal p-value | 0.0034305318 |
| FDR q-value | 0.54997504 |
| FWER p-Value | 0.958 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CNKSR1 | 41 | 3.229 | 0.0166 | No | ||
| 2 | CNP | 195 | 2.524 | 0.0230 | No | ||
| 3 | KLF12 | 314 | 2.309 | 0.0301 | No | ||
| 4 | IGFBP7 | 383 | 2.209 | 0.0392 | No | ||
| 5 | PABPC4 | 464 | 2.106 | 0.0472 | No | ||
| 6 | CD83 | 535 | 2.032 | 0.0552 | No | ||
| 7 | DRD2 | 692 | 1.891 | 0.0578 | No | ||
| 8 | YTHDF2 | 712 | 1.878 | 0.0677 | No | ||
| 9 | PTPRG | 924 | 1.737 | 0.0664 | No | ||
| 10 | LHX2 | 1001 | 1.700 | 0.0722 | No | ||
| 11 | MAN1C1 | 1240 | 1.594 | 0.0685 | No | ||
| 12 | SPAG9 | 1456 | 1.499 | 0.0656 | No | ||
| 13 | SCN5A | 1494 | 1.483 | 0.0723 | No | ||
| 14 | KCNK12 | 1531 | 1.469 | 0.0789 | No | ||
| 15 | PABPC1 | 1959 | 1.327 | 0.0635 | No | ||
| 16 | CLDN7 | 2011 | 1.311 | 0.0683 | No | ||
| 17 | HNRPDL | 2036 | 1.305 | 0.0746 | No | ||
| 18 | E2F3 | 2258 | 1.247 | 0.0699 | No | ||
| 19 | NEO1 | 2263 | 1.246 | 0.0769 | No | ||
| 20 | UST | 2560 | 1.167 | 0.0677 | No | ||
| 21 | NUP62 | 2671 | 1.140 | 0.0684 | No | ||
| 22 | IL4I1 | 2692 | 1.135 | 0.0739 | No | ||
| 23 | DSCAM | 2987 | 1.061 | 0.0641 | No | ||
| 24 | MAP2K2 | 3227 | 1.012 | 0.0571 | No | ||
| 25 | PIP5K2B | 3319 | 0.992 | 0.0579 | No | ||
| 26 | CALM1 | 3352 | 0.982 | 0.0619 | No | ||
| 27 | CTCF | 3448 | 0.965 | 0.0624 | No | ||
| 28 | MRVI1 | 3909 | 0.872 | 0.0425 | No | ||
| 29 | ZNF385 | 3923 | 0.868 | 0.0469 | No | ||
| 30 | ARID1A | 4054 | 0.842 | 0.0447 | No | ||
| 31 | UBE2H | 4271 | 0.802 | 0.0377 | No | ||
| 32 | NFATC3 | 4333 | 0.790 | 0.0390 | No | ||
| 33 | ARTN | 4342 | 0.788 | 0.0432 | No | ||
| 34 | THRAP1 | 4365 | 0.784 | 0.0465 | No | ||
| 35 | LDB1 | 4449 | 0.768 | 0.0465 | No | ||
| 36 | JUP | 4687 | 0.724 | 0.0379 | No | ||
| 37 | PTMA | 4759 | 0.712 | 0.0382 | No | ||
| 38 | PAK3 | 4763 | 0.711 | 0.0421 | No | ||
| 39 | NXPH4 | 4970 | 0.673 | 0.0349 | No | ||
| 40 | SPATA6 | 5166 | 0.638 | 0.0280 | No | ||
| 41 | UBE1C | 5265 | 0.622 | 0.0264 | No | ||
| 42 | ATP5H | 5434 | 0.591 | 0.0207 | No | ||
| 43 | KCNJ12 | 5435 | 0.590 | 0.0241 | No | ||
| 44 | NR3C1 | 5615 | 0.565 | 0.0177 | No | ||
| 45 | PDGFB | 5688 | 0.553 | 0.0170 | No | ||
| 46 | POU3F1 | 5709 | 0.550 | 0.0191 | No | ||
| 47 | PRDM16 | 6095 | 0.486 | 0.0011 | No | ||
| 48 | SOX9 | 6127 | 0.481 | 0.0022 | No | ||
| 49 | BTBD14B | 6137 | 0.479 | 0.0045 | No | ||
| 50 | WNT10A | 6302 | 0.455 | -0.0017 | No | ||
| 51 | NR2C2 | 6701 | 0.398 | -0.0210 | No | ||
| 52 | KCNQ4 | 6706 | 0.397 | -0.0189 | No | ||
| 53 | PLXNC1 | 6818 | 0.381 | -0.0227 | No | ||
| 54 | LIG3 | 6882 | 0.370 | -0.0239 | No | ||
| 55 | LHX6 | 6933 | 0.362 | -0.0245 | No | ||
| 56 | CDON | 6976 | 0.356 | -0.0247 | No | ||
| 57 | KLF13 | 7314 | 0.313 | -0.0412 | No | ||
| 58 | HTATIP | 7344 | 0.308 | -0.0410 | No | ||
| 59 | GMIP | 7414 | 0.299 | -0.0430 | No | ||
| 60 | POU3F2 | 7473 | 0.290 | -0.0444 | No | ||
| 61 | ARPC4 | 7580 | 0.274 | -0.0486 | No | ||
| 62 | ATP6V1D | 7601 | 0.270 | -0.0481 | No | ||
| 63 | APLP1 | 7675 | 0.259 | -0.0505 | No | ||
| 64 | RFXDC1 | 7836 | 0.231 | -0.0579 | No | ||
| 65 | LMO4 | 7980 | 0.207 | -0.0644 | No | ||
| 66 | MKNK2 | 8036 | 0.200 | -0.0662 | No | ||
| 67 | HOXC10 | 8335 | 0.155 | -0.0815 | No | ||
| 68 | FKBP14 | 8493 | 0.131 | -0.0892 | No | ||
| 69 | APLP2 | 8515 | 0.129 | -0.0896 | No | ||
| 70 | PTK7 | 8848 | 0.075 | -0.1072 | No | ||
| 71 | UBTF | 8980 | 0.050 | -0.1140 | No | ||
| 72 | FRMPD1 | 8982 | 0.049 | -0.1138 | No | ||
| 73 | LIN28 | 9081 | 0.032 | -0.1189 | No | ||
| 74 | NFIB | 9182 | 0.017 | -0.1242 | No | ||
| 75 | SP4 | 9244 | 0.009 | -0.1275 | No | ||
| 76 | ETV4 | 9365 | -0.012 | -0.1339 | No | ||
| 77 | TLK2 | 9791 | -0.074 | -0.1565 | No | ||
| 78 | DLL4 | 9949 | -0.098 | -0.1645 | No | ||
| 79 | EGLN2 | 10209 | -0.142 | -0.1777 | No | ||
| 80 | MAP3K6 | 10293 | -0.154 | -0.1813 | No | ||
| 81 | CENPB | 10347 | -0.165 | -0.1832 | No | ||
| 82 | DULLARD | 10466 | -0.183 | -0.1885 | No | ||
| 83 | SIX1 | 10474 | -0.184 | -0.1878 | No | ||
| 84 | RPIA | 10543 | -0.193 | -0.1904 | No | ||
| 85 | JMJD1B | 10620 | -0.205 | -0.1933 | No | ||
| 86 | FKRP | 10631 | -0.207 | -0.1927 | No | ||
| 87 | HTR7 | 10679 | -0.215 | -0.1940 | No | ||
| 88 | SLC12A4 | 10880 | -0.243 | -0.2034 | No | ||
| 89 | MAP3K11 | 11008 | -0.261 | -0.2088 | No | ||
| 90 | EIF3S10 | 11043 | -0.267 | -0.2091 | No | ||
| 91 | GDF8 | 11168 | -0.285 | -0.2141 | No | ||
| 92 | TRIM28 | 11233 | -0.295 | -0.2159 | No | ||
| 93 | SLC25A28 | 11262 | -0.299 | -0.2156 | No | ||
| 94 | TITF1 | 11427 | -0.328 | -0.2226 | No | ||
| 95 | RELB | 11504 | -0.340 | -0.2248 | No | ||
| 96 | CXXC5 | 11629 | -0.360 | -0.2294 | No | ||
| 97 | SCML2 | 11694 | -0.371 | -0.2307 | No | ||
| 98 | ERF | 11745 | -0.378 | -0.2312 | No | ||
| 99 | CDC14A | 11904 | -0.405 | -0.2374 | No | ||
| 100 | HPN | 12086 | -0.434 | -0.2447 | No | ||
| 101 | STAT3 | 12107 | -0.438 | -0.2432 | No | ||
| 102 | IL11 | 12223 | -0.455 | -0.2468 | No | ||
| 103 | SLC39A7 | 12413 | -0.488 | -0.2542 | No | ||
| 104 | UBE2M | 12864 | -0.564 | -0.2753 | No | ||
| 105 | THAP11 | 12969 | -0.580 | -0.2776 | No | ||
| 106 | PHC2 | 13091 | -0.600 | -0.2807 | No | ||
| 107 | CREB3L1 | 13133 | -0.607 | -0.2794 | No | ||
| 108 | HOXA1 | 13298 | -0.639 | -0.2845 | No | ||
| 109 | MAFB | 13358 | -0.647 | -0.2839 | No | ||
| 110 | GPRC5C | 13411 | -0.657 | -0.2829 | No | ||
| 111 | SMAD7 | 13703 | -0.711 | -0.2946 | No | ||
| 112 | DSCAML1 | 13714 | -0.713 | -0.2910 | No | ||
| 113 | CIC | 13852 | -0.737 | -0.2941 | No | ||
| 114 | HOXA3 | 13977 | -0.759 | -0.2964 | No | ||
| 115 | BARX1 | 13992 | -0.762 | -0.2927 | No | ||
| 116 | SIPA1 | 14069 | -0.779 | -0.2923 | No | ||
| 117 | TEAD2 | 14137 | -0.792 | -0.2913 | No | ||
| 118 | SEMA6C | 14139 | -0.792 | -0.2867 | No | ||
| 119 | RXRB | 14254 | -0.813 | -0.2882 | No | ||
| 120 | PRR3 | 14419 | -0.846 | -0.2921 | No | ||
| 121 | HIF1A | 14654 | -0.887 | -0.2997 | No | ||
| 122 | SOX5 | 14737 | -0.906 | -0.2988 | No | ||
| 123 | RARG | 14778 | -0.915 | -0.2957 | No | ||
| 124 | P4HA1 | 14856 | -0.932 | -0.2944 | No | ||
| 125 | BAT3 | 15143 | -0.997 | -0.3041 | No | ||
| 126 | SLC30A3 | 15154 | -0.999 | -0.2988 | No | ||
| 127 | GAS7 | 15509 | -1.081 | -0.3117 | Yes | ||
| 128 | MXD4 | 15542 | -1.092 | -0.3071 | Yes | ||
| 129 | FGF11 | 15551 | -1.094 | -0.3012 | Yes | ||
| 130 | ANKRD2 | 15560 | -1.096 | -0.2952 | Yes | ||
| 131 | MDS1 | 15614 | -1.109 | -0.2916 | Yes | ||
| 132 | CDK9 | 15634 | -1.113 | -0.2862 | Yes | ||
| 133 | ATF5 | 15959 | -1.202 | -0.2967 | Yes | ||
| 134 | KCNH8 | 16032 | -1.218 | -0.2935 | Yes | ||
| 135 | TRPC1 | 16034 | -1.219 | -0.2865 | Yes | ||
| 136 | JARID2 | 16108 | -1.238 | -0.2832 | Yes | ||
| 137 | TRERF1 | 16190 | -1.262 | -0.2803 | Yes | ||
| 138 | DGCR2 | 16292 | -1.292 | -0.2782 | Yes | ||
| 139 | DGKG | 16308 | -1.296 | -0.2715 | Yes | ||
| 140 | MFNG | 16351 | -1.310 | -0.2661 | Yes | ||
| 141 | HR | 16391 | -1.322 | -0.2605 | Yes | ||
| 142 | SIX4 | 16515 | -1.366 | -0.2593 | Yes | ||
| 143 | RASIP1 | 16611 | -1.395 | -0.2563 | Yes | ||
| 144 | EFNB3 | 16636 | -1.404 | -0.2494 | Yes | ||
| 145 | SLITRK5 | 16723 | -1.438 | -0.2457 | Yes | ||
| 146 | HNRPL | 16781 | -1.461 | -0.2403 | Yes | ||
| 147 | MYH7 | 16827 | -1.478 | -0.2341 | Yes | ||
| 148 | DPF3 | 17064 | -1.572 | -0.2377 | Yes | ||
| 149 | PITX2 | 17203 | -1.627 | -0.2357 | Yes | ||
| 150 | DAB1 | 17222 | -1.635 | -0.2272 | Yes | ||
| 151 | ROCK1 | 17278 | -1.656 | -0.2205 | Yes | ||
| 152 | SOS1 | 17388 | -1.701 | -0.2165 | Yes | ||
| 153 | SOX10 | 17406 | -1.709 | -0.2075 | Yes | ||
| 154 | LRP8 | 17412 | -1.711 | -0.1978 | Yes | ||
| 155 | PDLIM2 | 17418 | -1.712 | -0.1881 | Yes | ||
| 156 | ANGPTL2 | 17476 | -1.742 | -0.1810 | Yes | ||
| 157 | FBS1 | 17496 | -1.753 | -0.1718 | Yes | ||
| 158 | FBXO34 | 17542 | -1.773 | -0.1639 | Yes | ||
| 159 | GRIK3 | 17671 | -1.849 | -0.1601 | Yes | ||
| 160 | PLP2 | 17700 | -1.865 | -0.1508 | Yes | ||
| 161 | CIB1 | 17801 | -1.935 | -0.1449 | Yes | ||
| 162 | DTX3 | 17891 | -1.989 | -0.1381 | Yes | ||
| 163 | CHRDL1 | 17910 | -2.001 | -0.1275 | Yes | ||
| 164 | HIC1 | 17921 | -2.010 | -0.1163 | Yes | ||
| 165 | NFAT5 | 17923 | -2.010 | -0.1046 | Yes | ||
| 166 | EIF2S1 | 17933 | -2.016 | -0.0934 | Yes | ||
| 167 | TMEM24 | 17950 | -2.025 | -0.0824 | Yes | ||
| 168 | CABP7 | 17957 | -2.034 | -0.0709 | Yes | ||
| 169 | KITLG | 18005 | -2.078 | -0.0614 | Yes | ||
| 170 | CRYAB | 18090 | -2.162 | -0.0533 | Yes | ||
| 171 | CEECAM1 | 18254 | -2.354 | -0.0484 | Yes | ||
| 172 | HSPB2 | 18293 | -2.410 | -0.0365 | Yes | ||
| 173 | BCL3 | 18477 | -2.823 | -0.0299 | Yes | ||
| 174 | OBSCN | 18515 | -2.965 | -0.0147 | Yes | ||
| 175 | NOTCH3 | 18582 | -3.447 | 0.0018 | Yes |
