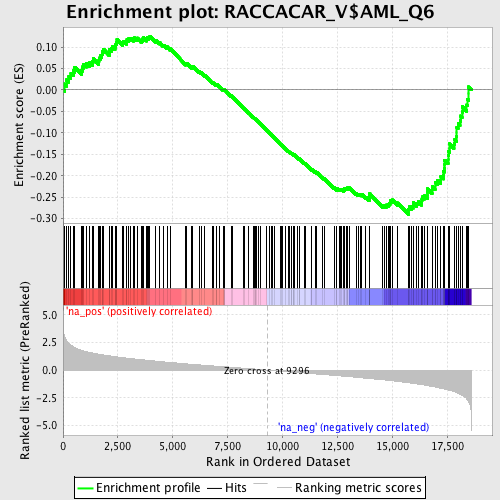
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RACCACAR_V$AML_Q6 |
| Enrichment Score (ES) | -0.2903308 |
| Normalized Enrichment Score (NES) | -1.4047745 |
| Nominal p-value | 0.015679443 |
| FDR q-value | 0.5340621 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HEMGN | 59 | 3.056 | 0.0139 | No | ||
| 2 | GTF2A1 | 158 | 2.667 | 0.0236 | No | ||
| 3 | EMP1 | 262 | 2.386 | 0.0313 | No | ||
| 4 | NEK6 | 356 | 2.236 | 0.0388 | No | ||
| 5 | GPX1 | 465 | 2.105 | 0.0448 | No | ||
| 6 | DAB2 | 529 | 2.043 | 0.0528 | No | ||
| 7 | CBL | 836 | 1.792 | 0.0462 | No | ||
| 8 | MSI2 | 882 | 1.764 | 0.0537 | No | ||
| 9 | FGFR1 | 937 | 1.731 | 0.0604 | No | ||
| 10 | KCTD15 | 1069 | 1.669 | 0.0627 | No | ||
| 11 | WNT8B | 1217 | 1.605 | 0.0637 | No | ||
| 12 | PPARG | 1339 | 1.551 | 0.0658 | No | ||
| 13 | TAGAP | 1365 | 1.537 | 0.0731 | No | ||
| 14 | BMPR1B | 1611 | 1.436 | 0.0679 | No | ||
| 15 | ATF7IP | 1636 | 1.430 | 0.0746 | No | ||
| 16 | RYBP | 1690 | 1.411 | 0.0796 | No | ||
| 17 | FOXP3 | 1772 | 1.383 | 0.0829 | No | ||
| 18 | LUC7L | 1784 | 1.380 | 0.0901 | No | ||
| 19 | PAFAH1B1 | 1832 | 1.367 | 0.0952 | No | ||
| 20 | TACC2 | 2101 | 1.286 | 0.0879 | No | ||
| 21 | BNC2 | 2122 | 1.280 | 0.0940 | No | ||
| 22 | CD28 | 2213 | 1.256 | 0.0961 | No | ||
| 23 | BATF | 2257 | 1.247 | 0.1008 | No | ||
| 24 | PSCDBP | 2366 | 1.218 | 0.1017 | No | ||
| 25 | CITED1 | 2402 | 1.206 | 0.1066 | No | ||
| 26 | IL17RE | 2431 | 1.199 | 0.1118 | No | ||
| 27 | CCL4 | 2454 | 1.194 | 0.1173 | No | ||
| 28 | POU2F1 | 2703 | 1.133 | 0.1102 | No | ||
| 29 | TCF7 | 2755 | 1.117 | 0.1137 | No | ||
| 30 | OLIG3 | 2884 | 1.082 | 0.1128 | No | ||
| 31 | ARHGAP8 | 2909 | 1.077 | 0.1175 | No | ||
| 32 | TCF12 | 2991 | 1.060 | 0.1191 | No | ||
| 33 | TBL1X | 3065 | 1.046 | 0.1210 | No | ||
| 34 | TBX5 | 3198 | 1.017 | 0.1195 | No | ||
| 35 | SLC36A2 | 3266 | 1.004 | 0.1215 | No | ||
| 36 | RGS1 | 3375 | 0.977 | 0.1211 | No | ||
| 37 | PDE6H | 3571 | 0.942 | 0.1158 | No | ||
| 38 | SCNN1A | 3612 | 0.935 | 0.1189 | No | ||
| 39 | MAFG | 3653 | 0.927 | 0.1219 | No | ||
| 40 | SLC31A2 | 3800 | 0.895 | 0.1190 | No | ||
| 41 | IFNG | 3840 | 0.886 | 0.1219 | No | ||
| 42 | RPA3 | 3907 | 0.872 | 0.1232 | No | ||
| 43 | CCL2 | 3954 | 0.862 | 0.1255 | No | ||
| 44 | CSF2 | 4215 | 0.810 | 0.1159 | No | ||
| 45 | HLX1 | 4380 | 0.781 | 0.1114 | No | ||
| 46 | ENTPD3 | 4587 | 0.741 | 0.1044 | No | ||
| 47 | BCL6 | 4734 | 0.715 | 0.1005 | No | ||
| 48 | RIN1 | 4890 | 0.689 | 0.0959 | No | ||
| 49 | PAX1 | 5591 | 0.569 | 0.0611 | No | ||
| 50 | GPD1 | 5634 | 0.561 | 0.0620 | No | ||
| 51 | LIF | 5844 | 0.525 | 0.0536 | No | ||
| 52 | CCR1 | 5901 | 0.518 | 0.0535 | No | ||
| 53 | AKT2 | 5918 | 0.515 | 0.0555 | No | ||
| 54 | TBPL1 | 6206 | 0.469 | 0.0426 | No | ||
| 55 | EDARADD | 6287 | 0.457 | 0.0408 | No | ||
| 56 | DOK2 | 6461 | 0.434 | 0.0338 | No | ||
| 57 | PLXNC1 | 6818 | 0.381 | 0.0167 | No | ||
| 58 | NDUFA3 | 6860 | 0.373 | 0.0165 | No | ||
| 59 | SCRN1 | 6974 | 0.356 | 0.0124 | No | ||
| 60 | HHIP | 7006 | 0.352 | 0.0127 | No | ||
| 61 | PDGFC | 7118 | 0.337 | 0.0085 | No | ||
| 62 | CD69 | 7317 | 0.313 | -0.0004 | No | ||
| 63 | WBP1 | 7343 | 0.308 | -0.0001 | No | ||
| 64 | SIRT1 | 7349 | 0.307 | 0.0014 | No | ||
| 65 | FCER1G | 7652 | 0.263 | -0.0135 | No | ||
| 66 | DMP1 | 7741 | 0.247 | -0.0169 | No | ||
| 67 | MATN1 | 8234 | 0.170 | -0.0427 | No | ||
| 68 | PICALM | 8280 | 0.162 | -0.0442 | No | ||
| 69 | PGF | 8468 | 0.134 | -0.0536 | No | ||
| 70 | CTHRC1 | 8657 | 0.109 | -0.0632 | No | ||
| 71 | PITPNC1 | 8732 | 0.096 | -0.0666 | No | ||
| 72 | IL3 | 8776 | 0.087 | -0.0685 | No | ||
| 73 | RUNX1 | 8777 | 0.087 | -0.0680 | No | ||
| 74 | CHAD | 8806 | 0.083 | -0.0691 | No | ||
| 75 | ETF1 | 8892 | 0.067 | -0.0733 | No | ||
| 76 | DYRK1B | 8995 | 0.047 | -0.0786 | No | ||
| 77 | HOXB5 | 9265 | 0.006 | -0.0931 | No | ||
| 78 | GABRA6 | 9383 | -0.014 | -0.0994 | No | ||
| 79 | MSRA | 9490 | -0.030 | -0.1050 | No | ||
| 80 | FLI1 | 9557 | -0.040 | -0.1083 | No | ||
| 81 | HOXD10 | 9611 | -0.048 | -0.1109 | No | ||
| 82 | EVI2A | 9908 | -0.091 | -0.1265 | No | ||
| 83 | ELAVL2 | 9967 | -0.101 | -0.1291 | No | ||
| 84 | THBS3 | 9991 | -0.105 | -0.1297 | No | ||
| 85 | OSCAR | 10153 | -0.132 | -0.1377 | No | ||
| 86 | PIK3R1 | 10260 | -0.150 | -0.1426 | No | ||
| 87 | ATBF1 | 10321 | -0.160 | -0.1450 | No | ||
| 88 | CALR | 10390 | -0.171 | -0.1477 | No | ||
| 89 | RGS14 | 10420 | -0.176 | -0.1483 | No | ||
| 90 | TRPM8 | 10494 | -0.187 | -0.1512 | No | ||
| 91 | CRY1 | 10497 | -0.187 | -0.1503 | No | ||
| 92 | PHF6 | 10528 | -0.191 | -0.1508 | No | ||
| 93 | AGTRL1 | 10676 | -0.214 | -0.1576 | No | ||
| 94 | SUPT4H1 | 10759 | -0.228 | -0.1608 | No | ||
| 95 | PROK2 | 10766 | -0.229 | -0.1598 | No | ||
| 96 | NAPSA | 10988 | -0.258 | -0.1703 | No | ||
| 97 | SUPT16H | 11032 | -0.265 | -0.1712 | No | ||
| 98 | RBBP6 | 11331 | -0.310 | -0.1856 | No | ||
| 99 | APOB48R | 11342 | -0.312 | -0.1844 | No | ||
| 100 | RASSF2 | 11483 | -0.337 | -0.1901 | No | ||
| 101 | AMD1 | 11561 | -0.350 | -0.1923 | No | ||
| 102 | HOXC4 | 11823 | -0.391 | -0.2043 | No | ||
| 103 | ATP5A1 | 11896 | -0.404 | -0.2059 | No | ||
| 104 | ABHD8 | 12350 | -0.477 | -0.2278 | No | ||
| 105 | PTPN22 | 12473 | -0.498 | -0.2317 | No | ||
| 106 | MTMR4 | 12482 | -0.499 | -0.2293 | No | ||
| 107 | BLOC1S1 | 12581 | -0.516 | -0.2317 | No | ||
| 108 | OTP | 12645 | -0.527 | -0.2322 | No | ||
| 109 | ADAMTS8 | 12691 | -0.536 | -0.2316 | No | ||
| 110 | LCK | 12782 | -0.552 | -0.2334 | No | ||
| 111 | MTMR3 | 12819 | -0.558 | -0.2322 | No | ||
| 112 | MAP1LC3A | 12829 | -0.560 | -0.2296 | No | ||
| 113 | SOST | 12896 | -0.570 | -0.2300 | No | ||
| 114 | IL7R | 12957 | -0.577 | -0.2300 | No | ||
| 115 | AKAP3 | 12977 | -0.582 | -0.2278 | No | ||
| 116 | SREBF2 | 13047 | -0.592 | -0.2282 | No | ||
| 117 | CPNE1 | 13356 | -0.647 | -0.2413 | No | ||
| 118 | TULP4 | 13464 | -0.666 | -0.2433 | No | ||
| 119 | RGMA | 13534 | -0.681 | -0.2433 | No | ||
| 120 | PCSK5 | 13609 | -0.694 | -0.2434 | No | ||
| 121 | KCNJ1 | 13802 | -0.729 | -0.2497 | No | ||
| 122 | CHES1 | 13953 | -0.755 | -0.2536 | No | ||
| 123 | AGPAT4 | 13963 | -0.758 | -0.2499 | No | ||
| 124 | COMMD5 | 13965 | -0.758 | -0.2457 | No | ||
| 125 | POF1B | 13979 | -0.759 | -0.2421 | No | ||
| 126 | PPP1R12C | 14576 | -0.873 | -0.2696 | No | ||
| 127 | NUCB2 | 14663 | -0.890 | -0.2692 | No | ||
| 128 | SOX5 | 14737 | -0.906 | -0.2681 | No | ||
| 129 | GPR50 | 14822 | -0.927 | -0.2675 | No | ||
| 130 | TLN1 | 14857 | -0.932 | -0.2641 | No | ||
| 131 | TNFRSF12A | 14901 | -0.941 | -0.2612 | No | ||
| 132 | POLG2 | 14933 | -0.950 | -0.2575 | No | ||
| 133 | ITGB7 | 15011 | -0.966 | -0.2563 | No | ||
| 134 | UBE2R2 | 15247 | -1.020 | -0.2633 | No | ||
| 135 | HOXB3 | 15746 | -1.143 | -0.2839 | Yes | ||
| 136 | DAB2IP | 15750 | -1.145 | -0.2777 | Yes | ||
| 137 | HOXA9 | 15773 | -1.151 | -0.2724 | Yes | ||
| 138 | ACIN1 | 15879 | -1.180 | -0.2715 | Yes | ||
| 139 | C1QTNF6 | 15950 | -1.201 | -0.2686 | Yes | ||
| 140 | HPCAL1 | 15967 | -1.204 | -0.2627 | Yes | ||
| 141 | CCNT2 | 16124 | -1.242 | -0.2642 | Yes | ||
| 142 | IL17F | 16188 | -1.261 | -0.2605 | Yes | ||
| 143 | PPP1R14C | 16337 | -1.307 | -0.2613 | Yes | ||
| 144 | ASB7 | 16352 | -1.310 | -0.2547 | Yes | ||
| 145 | SLC12A6 | 16390 | -1.322 | -0.2493 | Yes | ||
| 146 | LIFR | 16485 | -1.353 | -0.2468 | Yes | ||
| 147 | EIF5A | 16599 | -1.392 | -0.2451 | Yes | ||
| 148 | DKK1 | 16607 | -1.394 | -0.2377 | Yes | ||
| 149 | PTPRS | 16620 | -1.398 | -0.2305 | Yes | ||
| 150 | ARHGEF2 | 16817 | -1.472 | -0.2329 | Yes | ||
| 151 | LTBP1 | 16824 | -1.476 | -0.2249 | Yes | ||
| 152 | SDF2L1 | 16954 | -1.524 | -0.2234 | Yes | ||
| 153 | RHOG | 16955 | -1.524 | -0.2149 | Yes | ||
| 154 | ZIC4 | 17057 | -1.568 | -0.2116 | Yes | ||
| 155 | TRPV2 | 17197 | -1.623 | -0.2100 | Yes | ||
| 156 | ZBTB8 | 17218 | -1.634 | -0.2019 | Yes | ||
| 157 | MTX1 | 17324 | -1.676 | -0.1982 | Yes | ||
| 158 | DGKZ | 17338 | -1.680 | -0.1895 | Yes | ||
| 159 | CXCR3 | 17374 | -1.697 | -0.1819 | Yes | ||
| 160 | ERG | 17385 | -1.700 | -0.1729 | Yes | ||
| 161 | ADAM28 | 17392 | -1.702 | -0.1637 | Yes | ||
| 162 | MKLN1 | 17546 | -1.775 | -0.1621 | Yes | ||
| 163 | EYA1 | 17549 | -1.777 | -0.1522 | Yes | ||
| 164 | BCAR3 | 17574 | -1.793 | -0.1435 | Yes | ||
| 165 | NUTF2 | 17621 | -1.820 | -0.1358 | Yes | ||
| 166 | EIF2C1 | 17624 | -1.822 | -0.1257 | Yes | ||
| 167 | REL | 17816 | -1.941 | -0.1252 | Yes | ||
| 168 | AP1S2 | 17853 | -1.959 | -0.1161 | Yes | ||
| 169 | HOXB6 | 17935 | -2.018 | -0.1092 | Yes | ||
| 170 | SLC26A7 | 17945 | -2.022 | -0.0984 | Yes | ||
| 171 | HEY1 | 17946 | -2.023 | -0.0870 | Yes | ||
| 172 | HOXC6 | 18002 | -2.076 | -0.0784 | Yes | ||
| 173 | HOXB4 | 18091 | -2.164 | -0.0710 | Yes | ||
| 174 | RPP21 | 18127 | -2.196 | -0.0606 | Yes | ||
| 175 | RUNX2 | 18185 | -2.266 | -0.0510 | Yes | ||
| 176 | JMJD1C | 18198 | -2.283 | -0.0389 | Yes | ||
| 177 | MADD | 18396 | -2.586 | -0.0351 | Yes | ||
| 178 | RTN4RL1 | 18438 | -2.685 | -0.0223 | Yes | ||
| 179 | WAS | 18484 | -2.836 | -0.0088 | Yes | ||
| 180 | CD6 | 18485 | -2.841 | 0.0071 | Yes |
