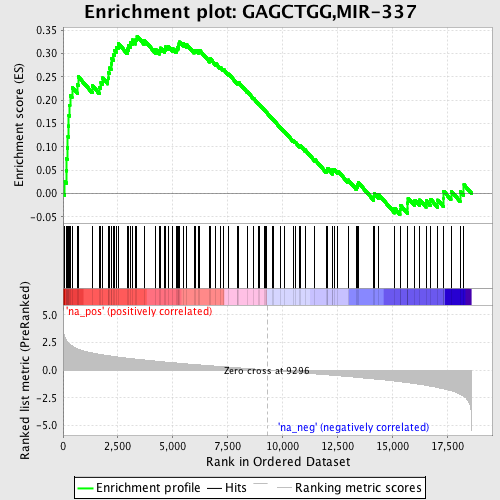
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GAGCTGG,MIR-337 |
| Enrichment Score (ES) | 0.33695897 |
| Normalized Enrichment Score (NES) | 1.5490117 |
| Nominal p-value | 0.011312217 |
| FDR q-value | 0.25741953 |
| FWER p-Value | 0.903 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SORT1 | 77 | 2.990 | 0.0256 | Yes | ||
| 2 | CD44 | 147 | 2.701 | 0.0487 | Yes | ||
| 3 | NR1D1 | 162 | 2.653 | 0.0744 | Yes | ||
| 4 | OVOL1 | 189 | 2.541 | 0.0983 | Yes | ||
| 5 | TOMM34 | 218 | 2.477 | 0.1214 | Yes | ||
| 6 | PACSIN2 | 236 | 2.427 | 0.1446 | Yes | ||
| 7 | NAV1 | 261 | 2.387 | 0.1671 | Yes | ||
| 8 | YPEL1 | 290 | 2.346 | 0.1889 | Yes | ||
| 9 | KLF12 | 314 | 2.309 | 0.2107 | Yes | ||
| 10 | H1F0 | 409 | 2.177 | 0.2272 | Yes | ||
| 11 | GLCCI1 | 650 | 1.918 | 0.2334 | Yes | ||
| 12 | EFNB1 | 689 | 1.892 | 0.2501 | Yes | ||
| 13 | UHRF2 | 1335 | 1.552 | 0.2307 | Yes | ||
| 14 | JPH1 | 1655 | 1.422 | 0.2276 | Yes | ||
| 15 | CHD4 | 1721 | 1.401 | 0.2381 | Yes | ||
| 16 | COMMD3 | 1801 | 1.377 | 0.2475 | Yes | ||
| 17 | ABCF1 | 2045 | 1.302 | 0.2473 | Yes | ||
| 18 | RAB15 | 2067 | 1.295 | 0.2591 | Yes | ||
| 19 | BNC2 | 2122 | 1.280 | 0.2689 | Yes | ||
| 20 | BTG2 | 2190 | 1.261 | 0.2778 | Yes | ||
| 21 | RNF44 | 2206 | 1.257 | 0.2895 | Yes | ||
| 22 | MFN1 | 2297 | 1.237 | 0.2970 | Yes | ||
| 23 | THRA | 2334 | 1.228 | 0.3072 | Yes | ||
| 24 | ZFAND3 | 2441 | 1.196 | 0.3134 | Yes | ||
| 25 | MAT2A | 2512 | 1.177 | 0.3213 | Yes | ||
| 26 | ACOT7 | 2928 | 1.072 | 0.3096 | Yes | ||
| 27 | DSCAM | 2987 | 1.061 | 0.3170 | Yes | ||
| 28 | ABCA9 | 3073 | 1.043 | 0.3228 | Yes | ||
| 29 | ZMYND11 | 3150 | 1.028 | 0.3289 | Yes | ||
| 30 | GPRASP2 | 3317 | 0.992 | 0.3298 | Yes | ||
| 31 | GRK6 | 3366 | 0.978 | 0.3370 | Yes | ||
| 32 | SEPT6 | 3693 | 0.917 | 0.3285 | No | ||
| 33 | SBK1 | 4216 | 0.809 | 0.3083 | No | ||
| 34 | BNIPL | 4399 | 0.775 | 0.3062 | No | ||
| 35 | AP1S3 | 4442 | 0.769 | 0.3116 | No | ||
| 36 | PARP6 | 4623 | 0.733 | 0.3092 | No | ||
| 37 | KCND1 | 4669 | 0.727 | 0.3140 | No | ||
| 38 | LPHN1 | 4782 | 0.709 | 0.3150 | No | ||
| 39 | SFRS1 | 4978 | 0.672 | 0.3111 | No | ||
| 40 | UBAP1 | 5162 | 0.640 | 0.3076 | No | ||
| 41 | MRPS16 | 5192 | 0.635 | 0.3123 | No | ||
| 42 | SLC1A2 | 5249 | 0.625 | 0.3155 | No | ||
| 43 | RASGRP4 | 5278 | 0.619 | 0.3202 | No | ||
| 44 | B4GALT3 | 5311 | 0.613 | 0.3246 | No | ||
| 45 | BRD2 | 5480 | 0.583 | 0.3213 | No | ||
| 46 | GPD1 | 5634 | 0.561 | 0.3186 | No | ||
| 47 | ITGA11 | 5966 | 0.506 | 0.3058 | No | ||
| 48 | TYSND1 | 6037 | 0.495 | 0.3069 | No | ||
| 49 | CHRD | 6158 | 0.476 | 0.3052 | No | ||
| 50 | SMARCD2 | 6218 | 0.467 | 0.3066 | No | ||
| 51 | SLC12A5 | 6688 | 0.399 | 0.2852 | No | ||
| 52 | DPH2 | 6700 | 0.398 | 0.2886 | No | ||
| 53 | RNF19 | 6967 | 0.357 | 0.2778 | No | ||
| 54 | SSBP3 | 7162 | 0.330 | 0.2706 | No | ||
| 55 | KLF13 | 7314 | 0.313 | 0.2655 | No | ||
| 56 | STIP1 | 7542 | 0.279 | 0.2561 | No | ||
| 57 | BCL2L1 | 7952 | 0.211 | 0.2361 | No | ||
| 58 | FOS | 7968 | 0.208 | 0.2373 | No | ||
| 59 | DNAJB5 | 7990 | 0.206 | 0.2382 | No | ||
| 60 | MAP3K3 | 8387 | 0.147 | 0.2183 | No | ||
| 61 | PARVA | 8685 | 0.103 | 0.2033 | No | ||
| 62 | MAP3K9 | 8882 | 0.068 | 0.1934 | No | ||
| 63 | PHF2 | 8938 | 0.057 | 0.1910 | No | ||
| 64 | TWIST1 | 9173 | 0.019 | 0.1785 | No | ||
| 65 | RGS16 | 9245 | 0.009 | 0.1748 | No | ||
| 66 | RARA | 9264 | 0.006 | 0.1739 | No | ||
| 67 | HOXB5 | 9265 | 0.006 | 0.1739 | No | ||
| 68 | CAMK2G | 9552 | -0.040 | 0.1589 | No | ||
| 69 | VCP | 9586 | -0.044 | 0.1575 | No | ||
| 70 | IER5L | 9929 | -0.095 | 0.1400 | No | ||
| 71 | FOSL1 | 10083 | -0.121 | 0.1329 | No | ||
| 72 | RANBP10 | 10491 | -0.187 | 0.1128 | No | ||
| 73 | SP7 | 10577 | -0.197 | 0.1102 | No | ||
| 74 | SEC24C | 10782 | -0.231 | 0.1014 | No | ||
| 75 | TXNL4B | 10808 | -0.235 | 0.1024 | No | ||
| 76 | KLF11 | 11025 | -0.264 | 0.0934 | No | ||
| 77 | POGZ | 11475 | -0.336 | 0.0725 | No | ||
| 78 | MCRS1 | 11994 | -0.420 | 0.0487 | No | ||
| 79 | ALDH1A2 | 12033 | -0.426 | 0.0508 | No | ||
| 80 | TEAD3 | 12053 | -0.429 | 0.0541 | No | ||
| 81 | PLAGL2 | 12290 | -0.467 | 0.0460 | No | ||
| 82 | BCAP29 | 12291 | -0.467 | 0.0506 | No | ||
| 83 | B3GAT3 | 12363 | -0.481 | 0.0516 | No | ||
| 84 | RNF121 | 12526 | -0.505 | 0.0479 | No | ||
| 85 | PPP1R3F | 12984 | -0.583 | 0.0290 | No | ||
| 86 | LIMK2 | 13375 | -0.650 | 0.0144 | No | ||
| 87 | EHMT2 | 13421 | -0.659 | 0.0185 | No | ||
| 88 | PHF13 | 13451 | -0.664 | 0.0235 | No | ||
| 89 | ROM1 | 14166 | -0.798 | -0.0071 | No | ||
| 90 | MAMDC1 | 14171 | -0.798 | 0.0006 | No | ||
| 91 | FBXL17 | 14381 | -0.837 | -0.0023 | No | ||
| 92 | LRRC4 | 15115 | -0.991 | -0.0321 | No | ||
| 93 | LBX1 | 15356 | -1.045 | -0.0346 | No | ||
| 94 | SLC25A22 | 15370 | -1.049 | -0.0249 | No | ||
| 95 | XBP1 | 15703 | -1.131 | -0.0316 | No | ||
| 96 | FRAG1 | 15704 | -1.131 | -0.0203 | No | ||
| 97 | JUNB | 15718 | -1.135 | -0.0097 | No | ||
| 98 | KCNH8 | 16032 | -1.218 | -0.0145 | No | ||
| 99 | TRIM35 | 16257 | -1.281 | -0.0139 | No | ||
| 100 | PDGFRB | 16542 | -1.375 | -0.0155 | No | ||
| 101 | TOR2A | 16751 | -1.449 | -0.0124 | No | ||
| 102 | PPP1R9B | 17073 | -1.577 | -0.0140 | No | ||
| 103 | SEMA6A | 17332 | -1.679 | -0.0112 | No | ||
| 104 | DGKZ | 17338 | -1.680 | 0.0052 | No | ||
| 105 | EI24 | 17681 | -1.855 | 0.0052 | No | ||
| 106 | VAMP1 | 18095 | -2.168 | 0.0044 | No | ||
| 107 | NFASC | 18269 | -2.376 | 0.0187 | No |
