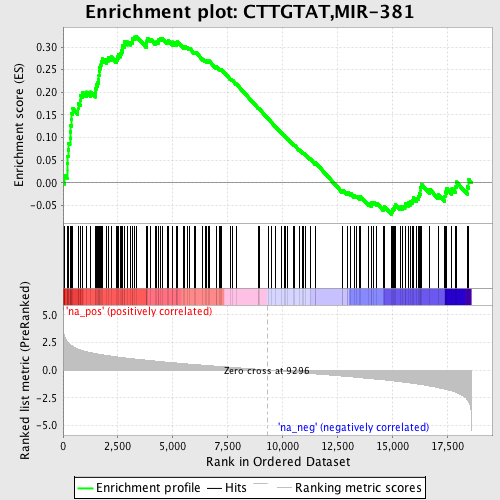
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CTTGTAT,MIR-381 |
| Enrichment Score (ES) | 0.32416338 |
| Normalized Enrichment Score (NES) | 1.5900149 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.39182246 |
| FWER p-Value | 0.795 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ANKRD50 | 55 | 3.082 | 0.0170 | Yes | ||
| 2 | SLMAP | 181 | 2.557 | 0.0267 | Yes | ||
| 3 | CEBPA | 182 | 2.555 | 0.0433 | Yes | ||
| 4 | GTF2I | 193 | 2.530 | 0.0591 | Yes | ||
| 5 | HIPK1 | 230 | 2.457 | 0.0730 | Yes | ||
| 6 | ZFYVE20 | 257 | 2.392 | 0.0871 | Yes | ||
| 7 | KLF12 | 314 | 2.309 | 0.0990 | Yes | ||
| 8 | KLF4 | 335 | 2.277 | 0.1126 | Yes | ||
| 9 | SEMA3C | 350 | 2.253 | 0.1264 | Yes | ||
| 10 | CXCR4 | 376 | 2.216 | 0.1394 | Yes | ||
| 11 | ELOVL7 | 386 | 2.203 | 0.1532 | Yes | ||
| 12 | GJA1 | 442 | 2.135 | 0.1640 | Yes | ||
| 13 | HBP1 | 678 | 1.898 | 0.1636 | Yes | ||
| 14 | MTSS1 | 687 | 1.892 | 0.1754 | Yes | ||
| 15 | DLC1 | 793 | 1.821 | 0.1815 | Yes | ||
| 16 | SYNCRIP | 800 | 1.816 | 0.1929 | Yes | ||
| 17 | RNF144 | 875 | 1.770 | 0.2003 | Yes | ||
| 18 | KCTD15 | 1069 | 1.669 | 0.2007 | Yes | ||
| 19 | STAG1 | 1259 | 1.586 | 0.2007 | Yes | ||
| 20 | BCL11B | 1468 | 1.494 | 0.1991 | Yes | ||
| 21 | CLCF1 | 1486 | 1.486 | 0.2078 | Yes | ||
| 22 | HNRPR | 1504 | 1.479 | 0.2164 | Yes | ||
| 23 | KPNA3 | 1583 | 1.448 | 0.2216 | Yes | ||
| 24 | NFKBIA | 1624 | 1.433 | 0.2287 | Yes | ||
| 25 | APPBP2 | 1631 | 1.431 | 0.2376 | Yes | ||
| 26 | COL3A1 | 1642 | 1.426 | 0.2463 | Yes | ||
| 27 | JPH1 | 1655 | 1.422 | 0.2548 | Yes | ||
| 28 | CHD4 | 1721 | 1.401 | 0.2604 | Yes | ||
| 29 | SEMA6D | 1732 | 1.398 | 0.2689 | Yes | ||
| 30 | BAT2 | 1802 | 1.376 | 0.2740 | Yes | ||
| 31 | ACSL3 | 1986 | 1.319 | 0.2727 | Yes | ||
| 32 | SLC24A3 | 2055 | 1.298 | 0.2774 | Yes | ||
| 33 | SLC38A2 | 2186 | 1.262 | 0.2785 | Yes | ||
| 34 | ZFAND3 | 2441 | 1.196 | 0.2725 | Yes | ||
| 35 | INSIG1 | 2476 | 1.187 | 0.2783 | Yes | ||
| 36 | UBE2E2 | 2530 | 1.174 | 0.2831 | Yes | ||
| 37 | EIF4G3 | 2626 | 1.152 | 0.2854 | Yes | ||
| 38 | SLC6A8 | 2645 | 1.148 | 0.2918 | Yes | ||
| 39 | SATB1 | 2705 | 1.132 | 0.2959 | Yes | ||
| 40 | EIF4G2 | 2713 | 1.129 | 0.3029 | Yes | ||
| 41 | KHDRBS1 | 2796 | 1.104 | 0.3056 | Yes | ||
| 42 | THRAP2 | 2806 | 1.102 | 0.3122 | Yes | ||
| 43 | SRRM1 | 2950 | 1.067 | 0.3114 | Yes | ||
| 44 | PDAP1 | 3081 | 1.041 | 0.3111 | Yes | ||
| 45 | ZMYND11 | 3150 | 1.028 | 0.3140 | Yes | ||
| 46 | FOXF2 | 3164 | 1.025 | 0.3199 | Yes | ||
| 47 | OSBPL3 | 3243 | 1.009 | 0.3223 | Yes | ||
| 48 | CSTF3 | 3327 | 0.990 | 0.3242 | Yes | ||
| 49 | PMP22 | 3805 | 0.893 | 0.3041 | No | ||
| 50 | RNF13 | 3811 | 0.892 | 0.3096 | No | ||
| 51 | EPHA3 | 3821 | 0.890 | 0.3149 | No | ||
| 52 | TBC1D15 | 3844 | 0.885 | 0.3194 | No | ||
| 53 | B3GNT5 | 3988 | 0.855 | 0.3172 | No | ||
| 54 | ZFPM2 | 4220 | 0.809 | 0.3099 | No | ||
| 55 | ARID4B | 4275 | 0.801 | 0.3122 | No | ||
| 56 | PNRC1 | 4340 | 0.788 | 0.3138 | No | ||
| 57 | THRAP1 | 4365 | 0.784 | 0.3176 | No | ||
| 58 | WEE1 | 4431 | 0.770 | 0.3190 | No | ||
| 59 | R3HDM1 | 4524 | 0.750 | 0.3189 | No | ||
| 60 | CNOT7 | 4743 | 0.713 | 0.3117 | No | ||
| 61 | LPHN1 | 4782 | 0.709 | 0.3142 | No | ||
| 62 | GAD1 | 4973 | 0.673 | 0.3083 | No | ||
| 63 | KCTD3 | 4999 | 0.667 | 0.3113 | No | ||
| 64 | BIRC6 | 5160 | 0.640 | 0.3067 | No | ||
| 65 | FLRT3 | 5176 | 0.637 | 0.3101 | No | ||
| 66 | RNF111 | 5225 | 0.628 | 0.3115 | No | ||
| 67 | ETS1 | 5492 | 0.581 | 0.3009 | No | ||
| 68 | RPS6KB1 | 5537 | 0.575 | 0.3022 | No | ||
| 69 | TRIM63 | 5649 | 0.559 | 0.2998 | No | ||
| 70 | BTBD7 | 5764 | 0.540 | 0.2971 | No | ||
| 71 | KIF1B | 5984 | 0.503 | 0.2885 | No | ||
| 72 | PAPOLA | 6046 | 0.494 | 0.2884 | No | ||
| 73 | APRIN | 6372 | 0.447 | 0.2737 | No | ||
| 74 | BHLHB5 | 6484 | 0.431 | 0.2705 | No | ||
| 75 | ABCA1 | 6557 | 0.419 | 0.2693 | No | ||
| 76 | ASXL2 | 6635 | 0.407 | 0.2678 | No | ||
| 77 | RAP2C | 6656 | 0.403 | 0.2693 | No | ||
| 78 | ZCCHC11 | 6970 | 0.356 | 0.2547 | No | ||
| 79 | GRM1 | 6972 | 0.356 | 0.2569 | No | ||
| 80 | PDGFC | 7118 | 0.337 | 0.2512 | No | ||
| 81 | EIF1 | 7164 | 0.330 | 0.2509 | No | ||
| 82 | RNF2 | 7216 | 0.324 | 0.2503 | No | ||
| 83 | PARD6B | 7641 | 0.264 | 0.2290 | No | ||
| 84 | UBE4A | 7716 | 0.253 | 0.2266 | No | ||
| 85 | SOX4 | 7894 | 0.220 | 0.2185 | No | ||
| 86 | ETF1 | 8892 | 0.067 | 0.1649 | No | ||
| 87 | RPL35A | 8921 | 0.062 | 0.1638 | No | ||
| 88 | PHF2 | 8938 | 0.057 | 0.1633 | No | ||
| 89 | MITF | 9345 | -0.008 | 0.1414 | No | ||
| 90 | ST8SIA2 | 9504 | -0.032 | 0.1330 | No | ||
| 91 | JAG2 | 9673 | -0.056 | 0.1243 | No | ||
| 92 | UBR1 | 9930 | -0.095 | 0.1110 | No | ||
| 93 | SSH2 | 10073 | -0.119 | 0.1041 | No | ||
| 94 | RHOQ | 10126 | -0.129 | 0.1021 | No | ||
| 95 | NAP1L5 | 10238 | -0.147 | 0.0971 | No | ||
| 96 | ITCH | 10506 | -0.188 | 0.0838 | No | ||
| 97 | ACVR2A | 10541 | -0.193 | 0.0832 | No | ||
| 98 | AZIN1 | 10767 | -0.229 | 0.0725 | No | ||
| 99 | LZTS2 | 10913 | -0.247 | 0.0663 | No | ||
| 100 | CGGBP1 | 10960 | -0.255 | 0.0654 | No | ||
| 101 | EIF3S10 | 11043 | -0.267 | 0.0627 | No | ||
| 102 | RPS6KA3 | 11264 | -0.299 | 0.0528 | No | ||
| 103 | TMEM16D | 11498 | -0.339 | 0.0423 | No | ||
| 104 | SHANK2 | 11500 | -0.340 | 0.0445 | No | ||
| 105 | PBEF1 | 12730 | -0.543 | -0.0186 | No | ||
| 106 | SNRK | 12753 | -0.547 | -0.0162 | No | ||
| 107 | VSNL1 | 12951 | -0.577 | -0.0231 | No | ||
| 108 | GFRA1 | 12978 | -0.582 | -0.0208 | No | ||
| 109 | DDX6 | 13119 | -0.603 | -0.0245 | No | ||
| 110 | NLK | 13271 | -0.634 | -0.0285 | No | ||
| 111 | PSIP1 | 13367 | -0.649 | -0.0295 | No | ||
| 112 | RAB11FIP2 | 13515 | -0.676 | -0.0331 | No | ||
| 113 | NR5A2 | 13558 | -0.686 | -0.0309 | No | ||
| 114 | NIN | 13919 | -0.748 | -0.0456 | No | ||
| 115 | YTHDF1 | 14064 | -0.779 | -0.0483 | No | ||
| 116 | DHX40 | 14071 | -0.779 | -0.0436 | No | ||
| 117 | XPO7 | 14164 | -0.797 | -0.0434 | No | ||
| 118 | BCL11A | 14288 | -0.819 | -0.0448 | No | ||
| 119 | NFIA | 14603 | -0.878 | -0.0561 | No | ||
| 120 | NDUFAB1 | 14646 | -0.885 | -0.0527 | No | ||
| 121 | QKI | 14970 | -0.957 | -0.0640 | No | ||
| 122 | PHF15 | 14995 | -0.962 | -0.0590 | No | ||
| 123 | EPHA4 | 15053 | -0.974 | -0.0558 | No | ||
| 124 | LRRC4 | 15115 | -0.991 | -0.0527 | No | ||
| 125 | BRD7 | 15163 | -1.001 | -0.0488 | No | ||
| 126 | ATRN | 15374 | -1.050 | -0.0534 | No | ||
| 127 | GRM8 | 15472 | -1.072 | -0.0517 | No | ||
| 128 | CRSP2 | 15583 | -1.100 | -0.0505 | No | ||
| 129 | MDS1 | 15614 | -1.109 | -0.0450 | No | ||
| 130 | ROD1 | 15738 | -1.140 | -0.0443 | No | ||
| 131 | GAS2 | 15834 | -1.167 | -0.0419 | No | ||
| 132 | UBE3A | 15912 | -1.190 | -0.0383 | No | ||
| 133 | AGPAT3 | 15956 | -1.202 | -0.0329 | No | ||
| 134 | CCNT2 | 16124 | -1.242 | -0.0339 | No | ||
| 135 | WNT5A | 16191 | -1.262 | -0.0293 | No | ||
| 136 | ACTG1 | 16232 | -1.274 | -0.0232 | No | ||
| 137 | MBNL2 | 16284 | -1.288 | -0.0177 | No | ||
| 138 | GORASP2 | 16288 | -1.290 | -0.0095 | No | ||
| 139 | ROCK2 | 16323 | -1.303 | -0.0029 | No | ||
| 140 | UBN1 | 16711 | -1.431 | -0.0146 | No | ||
| 141 | PURG | 17093 | -1.585 | -0.0250 | No | ||
| 142 | STX12 | 17395 | -1.704 | -0.0303 | No | ||
| 143 | HNRPH2 | 17414 | -1.712 | -0.0202 | No | ||
| 144 | PAPOLG | 17475 | -1.741 | -0.0122 | No | ||
| 145 | RBM16 | 17721 | -1.876 | -0.0133 | No | ||
| 146 | SIPA1L2 | 17884 | -1.986 | -0.0092 | No | ||
| 147 | NRXN1 | 17919 | -2.006 | 0.0019 | No | ||
| 148 | CACNA1C | 18420 | -2.639 | -0.0081 | No | ||
| 149 | RALGPS1 | 18493 | -2.882 | 0.0067 | No |
