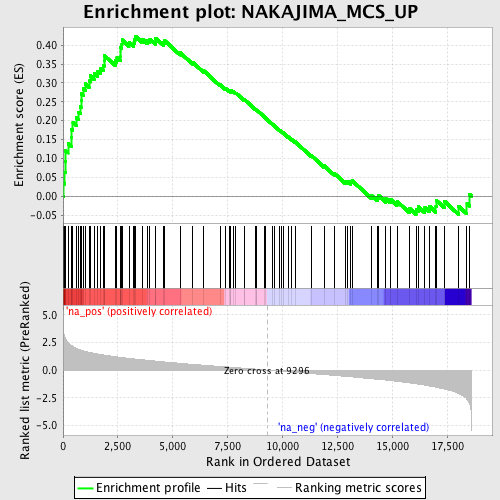
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | NAKAJIMA_MCS_UP |
| Enrichment Score (ES) | 0.4240998 |
| Normalized Enrichment Score (NES) | 1.9060622 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09166285 |
| FWER p-Value | 0.555 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL6R | 30 | 3.422 | 0.0339 | Yes | ||
| 2 | IL11RA1 | 67 | 3.026 | 0.0634 | Yes | ||
| 3 | SMOX | 96 | 2.892 | 0.0919 | Yes | ||
| 4 | CCL5 | 116 | 2.806 | 0.1200 | Yes | ||
| 5 | CX3CR1 | 223 | 2.463 | 0.1399 | Yes | ||
| 6 | CXCR4 | 376 | 2.216 | 0.1547 | Yes | ||
| 7 | CD63 | 382 | 2.209 | 0.1774 | Yes | ||
| 8 | LGALS3 | 448 | 2.122 | 0.1959 | Yes | ||
| 9 | CSF3 | 603 | 1.966 | 0.2080 | Yes | ||
| 10 | IL4R | 691 | 1.891 | 0.2229 | Yes | ||
| 11 | SPP1 | 781 | 1.828 | 0.2371 | Yes | ||
| 12 | ERO1L | 821 | 1.802 | 0.2537 | Yes | ||
| 13 | CCR4 | 827 | 1.799 | 0.2721 | Yes | ||
| 14 | CSF1 | 932 | 1.733 | 0.2845 | Yes | ||
| 15 | IL18 | 1015 | 1.693 | 0.2977 | Yes | ||
| 16 | CSF2RB1 | 1185 | 1.617 | 0.3053 | Yes | ||
| 17 | CSF3R | 1239 | 1.595 | 0.3190 | Yes | ||
| 18 | CXCL5 | 1413 | 1.519 | 0.3255 | Yes | ||
| 19 | CSF2RB2 | 1585 | 1.448 | 0.3313 | Yes | ||
| 20 | CLU | 1718 | 1.402 | 0.3387 | Yes | ||
| 21 | CCL7 | 1830 | 1.368 | 0.3469 | Yes | ||
| 22 | CSF1R | 1871 | 1.356 | 0.3588 | Yes | ||
| 23 | IL2 | 1881 | 1.353 | 0.3724 | Yes | ||
| 24 | HPGD | 2373 | 1.216 | 0.3585 | Yes | ||
| 25 | CCL4 | 2454 | 1.194 | 0.3666 | Yes | ||
| 26 | IL2RG | 2611 | 1.157 | 0.3702 | Yes | ||
| 27 | CX3CL1 | 2615 | 1.156 | 0.3820 | Yes | ||
| 28 | CAPG | 2616 | 1.156 | 0.3940 | Yes | ||
| 29 | GADD45B | 2678 | 1.139 | 0.4026 | Yes | ||
| 30 | CCL3 | 2684 | 1.138 | 0.4141 | Yes | ||
| 31 | IL1A | 3004 | 1.058 | 0.4079 | Yes | ||
| 32 | IFNGR1 | 3229 | 1.012 | 0.4063 | Yes | ||
| 33 | HNRPA1 | 3255 | 1.006 | 0.4154 | Yes | ||
| 34 | PF4 | 3287 | 0.999 | 0.4241 | Yes | ||
| 35 | TPH1 | 3620 | 0.932 | 0.4159 | No | ||
| 36 | IFNG | 3840 | 0.886 | 0.4132 | No | ||
| 37 | CCL2 | 3954 | 0.862 | 0.4161 | No | ||
| 38 | CSF2 | 4215 | 0.810 | 0.4105 | No | ||
| 39 | IL10RA | 4219 | 0.809 | 0.4187 | No | ||
| 40 | CCL8 | 4572 | 0.742 | 0.4074 | No | ||
| 41 | IL10 | 4621 | 0.733 | 0.4124 | No | ||
| 42 | IFNB1 | 5334 | 0.610 | 0.3803 | No | ||
| 43 | CCR1 | 5901 | 0.518 | 0.3552 | No | ||
| 44 | KIT | 6419 | 0.440 | 0.3318 | No | ||
| 45 | TIMP1 | 7157 | 0.331 | 0.2955 | No | ||
| 46 | CCR2 | 7395 | 0.302 | 0.2858 | No | ||
| 47 | IL1B | 7564 | 0.276 | 0.2796 | No | ||
| 48 | CYP11A1 | 7639 | 0.264 | 0.2784 | No | ||
| 49 | HIPK2 | 7645 | 0.263 | 0.2808 | No | ||
| 50 | IL9R | 7750 | 0.245 | 0.2778 | No | ||
| 51 | IL17A | 7877 | 0.223 | 0.2733 | No | ||
| 52 | CCL1 | 8258 | 0.166 | 0.2545 | No | ||
| 53 | IL6 | 8260 | 0.164 | 0.2561 | No | ||
| 54 | IL3 | 8776 | 0.087 | 0.2293 | No | ||
| 55 | IL7 | 8816 | 0.081 | 0.2280 | No | ||
| 56 | IL3RA | 8831 | 0.079 | 0.2281 | No | ||
| 57 | LOC242517 | 9189 | 0.017 | 0.2090 | No | ||
| 58 | IGFBP5 | 9208 | 0.014 | 0.2081 | No | ||
| 59 | CTSG | 9537 | -0.037 | 0.1908 | No | ||
| 60 | IL17RA | 9649 | -0.053 | 0.1854 | No | ||
| 61 | IL13 | 9880 | -0.087 | 0.1739 | No | ||
| 62 | CCL11 | 9936 | -0.096 | 0.1719 | No | ||
| 63 | CCR9 | 10036 | -0.112 | 0.1677 | No | ||
| 64 | IER3 | 10263 | -0.150 | 0.1571 | No | ||
| 65 | CPA3 | 10419 | -0.176 | 0.1505 | No | ||
| 66 | CCR3 | 10581 | -0.197 | 0.1439 | No | ||
| 67 | GEM | 11303 | -0.306 | 0.1082 | No | ||
| 68 | IL12B | 11890 | -0.404 | 0.0807 | No | ||
| 69 | IL2RB | 12357 | -0.479 | 0.0605 | No | ||
| 70 | LGALS1 | 12849 | -0.562 | 0.0399 | No | ||
| 71 | IL7R | 12957 | -0.577 | 0.0401 | No | ||
| 72 | HSP90AB1 | 13112 | -0.602 | 0.0380 | No | ||
| 73 | CRTAM | 13170 | -0.614 | 0.0413 | No | ||
| 74 | IL1R1 | 14074 | -0.780 | 0.0007 | No | ||
| 75 | IL12A | 14319 | -0.826 | -0.0039 | No | ||
| 76 | TNF | 14375 | -0.836 | 0.0018 | No | ||
| 77 | CCL22 | 14708 | -0.899 | -0.0068 | No | ||
| 78 | CCL25 | 14917 | -0.945 | -0.0082 | No | ||
| 79 | IL5 | 15218 | -1.015 | -0.0138 | No | ||
| 80 | FCER1A | 15779 | -1.152 | -0.0321 | No | ||
| 81 | ASNS | 16086 | -1.234 | -0.0358 | No | ||
| 82 | MCPT6 | 16187 | -1.261 | -0.0281 | No | ||
| 83 | CCR6 | 16490 | -1.356 | -0.0303 | No | ||
| 84 | HDC | 16693 | -1.425 | -0.0264 | No | ||
| 85 | TNFRSF9 | 16992 | -1.539 | -0.0265 | No | ||
| 86 | MAOA | 17017 | -1.553 | -0.0117 | No | ||
| 87 | CXCR3 | 17374 | -1.697 | -0.0133 | No | ||
| 88 | IL16 | 18025 | -2.097 | -0.0266 | No | ||
| 89 | IL2RA | 18407 | -2.610 | -0.0201 | No | ||
| 90 | IL1RL1 | 18524 | -3.015 | 0.0050 | No |
