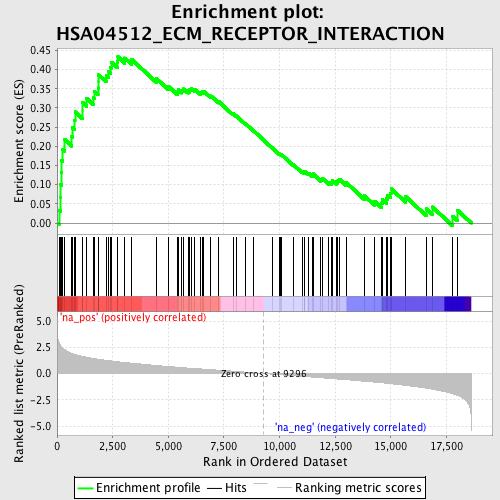
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HSA04512_ECM_RECEPTOR_INTERACTION |
| Enrichment Score (ES) | 0.43317536 |
| Normalized Enrichment Score (NES) | 1.908438 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.097602405 |
| FWER p-Value | 0.544 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SDC4 | 86 | 2.957 | 0.0340 | Yes | ||
| 2 | CD44 | 147 | 2.701 | 0.0660 | Yes | ||
| 3 | ITGB1 | 151 | 2.685 | 0.1009 | Yes | ||
| 4 | ITGB5 | 194 | 2.526 | 0.1316 | Yes | ||
| 5 | ITGA7 | 204 | 2.509 | 0.1639 | Yes | ||
| 6 | ITGA10 | 259 | 2.389 | 0.1922 | Yes | ||
| 7 | HSPG2 | 346 | 2.263 | 0.2171 | Yes | ||
| 8 | COL4A6 | 654 | 1.917 | 0.2255 | Yes | ||
| 9 | LAMC2 | 686 | 1.892 | 0.2486 | Yes | ||
| 10 | SPP1 | 781 | 1.828 | 0.2674 | Yes | ||
| 11 | GP6 | 806 | 1.810 | 0.2897 | Yes | ||
| 12 | TNC | 1146 | 1.631 | 0.2927 | Yes | ||
| 13 | THBS2 | 1147 | 1.630 | 0.3140 | Yes | ||
| 14 | FN1 | 1326 | 1.556 | 0.3247 | Yes | ||
| 15 | COL3A1 | 1642 | 1.426 | 0.3263 | Yes | ||
| 16 | GP1BB | 1666 | 1.418 | 0.3436 | Yes | ||
| 17 | TNN | 1840 | 1.365 | 0.3521 | Yes | ||
| 18 | LAMC3 | 1866 | 1.357 | 0.3685 | Yes | ||
| 19 | SDC3 | 1868 | 1.357 | 0.3861 | Yes | ||
| 20 | ITGAV | 2204 | 1.257 | 0.3845 | Yes | ||
| 21 | HMMR | 2315 | 1.233 | 0.3946 | Yes | ||
| 22 | ITGB4 | 2397 | 1.207 | 0.4060 | Yes | ||
| 23 | LAMA2 | 2437 | 1.197 | 0.4196 | Yes | ||
| 24 | LAMB2 | 2702 | 1.133 | 0.4201 | Yes | ||
| 25 | COL1A2 | 2733 | 1.124 | 0.4332 | Yes | ||
| 26 | RELN | 3043 | 1.050 | 0.4302 | No | ||
| 27 | COL4A1 | 3354 | 0.981 | 0.4263 | No | ||
| 28 | LAMC1 | 4456 | 0.765 | 0.3769 | No | ||
| 29 | IBSP | 5012 | 0.666 | 0.3557 | No | ||
| 30 | COL2A1 | 5414 | 0.594 | 0.3418 | No | ||
| 31 | CD47 | 5452 | 0.587 | 0.3475 | No | ||
| 32 | ITGA5 | 5612 | 0.565 | 0.3463 | No | ||
| 33 | COL6A3 | 5685 | 0.553 | 0.3496 | No | ||
| 34 | SDC1 | 5886 | 0.519 | 0.3456 | No | ||
| 35 | ITGA11 | 5966 | 0.506 | 0.3479 | No | ||
| 36 | ITGB6 | 6031 | 0.496 | 0.3510 | No | ||
| 37 | LAMA4 | 6182 | 0.472 | 0.3490 | No | ||
| 38 | ITGA6 | 6435 | 0.438 | 0.3412 | No | ||
| 39 | COL4A2 | 6520 | 0.426 | 0.3422 | No | ||
| 40 | COL1A1 | 6590 | 0.413 | 0.3438 | No | ||
| 41 | GP9 | 6912 | 0.365 | 0.3313 | No | ||
| 42 | FNDC4 | 7255 | 0.321 | 0.3170 | No | ||
| 43 | VTN | 7906 | 0.218 | 0.2848 | No | ||
| 44 | ITGA8 | 8060 | 0.195 | 0.2791 | No | ||
| 45 | COL5A1 | 8454 | 0.137 | 0.2597 | No | ||
| 46 | CHAD | 8806 | 0.083 | 0.2419 | No | ||
| 47 | ITGA2B | 9696 | -0.059 | 0.1947 | No | ||
| 48 | THBS3 | 9991 | -0.105 | 0.1802 | No | ||
| 49 | COL5A2 | 10046 | -0.114 | 0.1788 | No | ||
| 50 | COL5A3 | 10074 | -0.119 | 0.1789 | No | ||
| 51 | SDC2 | 10605 | -0.203 | 0.1529 | No | ||
| 52 | THBS4 | 11022 | -0.263 | 0.1339 | No | ||
| 53 | LAMA5 | 11113 | -0.278 | 0.1327 | No | ||
| 54 | GP1BA | 11136 | -0.280 | 0.1352 | No | ||
| 55 | SV2B | 11301 | -0.306 | 0.1303 | No | ||
| 56 | COL6A1 | 11484 | -0.337 | 0.1249 | No | ||
| 57 | SV2C | 11507 | -0.341 | 0.1282 | No | ||
| 58 | ITGB3 | 11860 | -0.399 | 0.1144 | No | ||
| 59 | ITGA3 | 11933 | -0.410 | 0.1159 | No | ||
| 60 | VWF | 12219 | -0.455 | 0.1064 | No | ||
| 61 | CD36 | 12354 | -0.478 | 0.1054 | No | ||
| 62 | COL6A2 | 12375 | -0.483 | 0.1107 | No | ||
| 63 | COL11A1 | 12547 | -0.509 | 0.1081 | No | ||
| 64 | COL11A2 | 12622 | -0.524 | 0.1109 | No | ||
| 65 | DAG1 | 12686 | -0.535 | 0.1145 | No | ||
| 66 | LAMA1 | 12992 | -0.584 | 0.1057 | No | ||
| 67 | ITGA4 | 13801 | -0.729 | 0.0716 | No | ||
| 68 | LAMA3 | 14265 | -0.815 | 0.0573 | No | ||
| 69 | TNXB | 14570 | -0.872 | 0.0523 | No | ||
| 70 | AGRN | 14617 | -0.880 | 0.0613 | No | ||
| 71 | LAMB3 | 14798 | -0.920 | 0.0636 | No | ||
| 72 | THBS1 | 14853 | -0.931 | 0.0728 | No | ||
| 73 | SV2A | 14988 | -0.961 | 0.0782 | No | ||
| 74 | ITGB7 | 15011 | -0.966 | 0.0896 | No | ||
| 75 | TNR | 15677 | -1.124 | 0.0684 | No | ||
| 76 | FNDC3A | 16597 | -1.391 | 0.0370 | No | ||
| 77 | COL4A4 | 16868 | -1.494 | 0.0419 | No | ||
| 78 | GP5 | 17787 | -1.923 | 0.0175 | No | ||
| 79 | ITGA9 | 18008 | -2.081 | 0.0328 | No |
