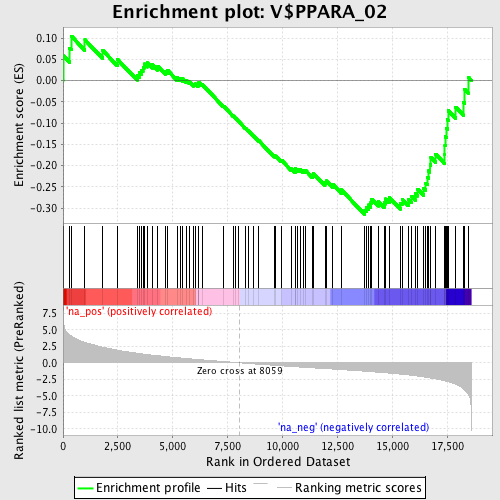
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$PPARA_02 |
| Enrichment Score (ES) | -0.31469423 |
| Normalized Enrichment Score (NES) | -1.3224931 |
| Nominal p-value | 0.069418386 |
| FDR q-value | 0.53016925 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DNAJC7 | 2 | 7.380 | 0.0588 | No | ||
| 2 | RTN4 | 299 | 4.226 | 0.0765 | No | ||
| 3 | KPNB1 | 374 | 4.033 | 0.1047 | No | ||
| 4 | CENTA2 | 995 | 3.084 | 0.0958 | No | ||
| 5 | HIP2 | 1808 | 2.357 | 0.0708 | No | ||
| 6 | RIN1 | 2483 | 1.905 | 0.0496 | No | ||
| 7 | PAK1 | 3380 | 1.444 | 0.0128 | No | ||
| 8 | HR | 3468 | 1.401 | 0.0193 | No | ||
| 9 | DHH | 3580 | 1.355 | 0.0241 | No | ||
| 10 | POGZ | 3644 | 1.319 | 0.0312 | No | ||
| 11 | SUPT16H | 3689 | 1.304 | 0.0393 | No | ||
| 12 | KCNB1 | 3825 | 1.244 | 0.0419 | No | ||
| 13 | NLGN3 | 4059 | 1.160 | 0.0386 | No | ||
| 14 | WDR20 | 4317 | 1.064 | 0.0332 | No | ||
| 15 | PLAGL2 | 4673 | 0.940 | 0.0215 | No | ||
| 16 | CHD2 | 4771 | 0.907 | 0.0235 | No | ||
| 17 | LOXL4 | 5196 | 0.775 | 0.0068 | No | ||
| 18 | RAD23B | 5328 | 0.734 | 0.0056 | No | ||
| 19 | ESRRG | 5457 | 0.691 | 0.0042 | No | ||
| 20 | RTN2 | 5638 | 0.635 | -0.0004 | No | ||
| 21 | TSNAXIP1 | 5775 | 0.600 | -0.0030 | No | ||
| 22 | HRH3 | 5960 | 0.549 | -0.0085 | No | ||
| 23 | FSCN3 | 6025 | 0.529 | -0.0077 | No | ||
| 24 | PHF12 | 6151 | 0.493 | -0.0105 | No | ||
| 25 | NEUROD2 | 6169 | 0.487 | -0.0076 | No | ||
| 26 | SEMA3A | 6185 | 0.484 | -0.0045 | No | ||
| 27 | UCN2 | 6331 | 0.444 | -0.0088 | No | ||
| 28 | CDK5R2 | 7304 | 0.191 | -0.0597 | No | ||
| 29 | CDH17 | 7746 | 0.079 | -0.0829 | No | ||
| 30 | WNT4 | 7865 | 0.050 | -0.0889 | No | ||
| 31 | LAMA5 | 8013 | 0.011 | -0.0967 | No | ||
| 32 | HMGN2 | 8310 | -0.061 | -0.1122 | No | ||
| 33 | OTX1 | 8430 | -0.095 | -0.1179 | No | ||
| 34 | RFX1 | 8669 | -0.154 | -0.1295 | No | ||
| 35 | SMARCA5 | 8915 | -0.212 | -0.1410 | No | ||
| 36 | LMO1 | 9634 | -0.374 | -0.1768 | No | ||
| 37 | TRPV6 | 9697 | -0.389 | -0.1770 | No | ||
| 38 | PURA | 9948 | -0.451 | -0.1869 | No | ||
| 39 | CYC1 | 10409 | -0.554 | -0.2073 | No | ||
| 40 | PDGFB | 10575 | -0.591 | -0.2115 | No | ||
| 41 | MAN2B1 | 10581 | -0.592 | -0.2070 | No | ||
| 42 | ZSWIM3 | 10690 | -0.617 | -0.2079 | No | ||
| 43 | MAFB | 10801 | -0.643 | -0.2087 | No | ||
| 44 | CACNA1S | 10953 | -0.672 | -0.2115 | No | ||
| 45 | CREB3L2 | 11031 | -0.687 | -0.2102 | No | ||
| 46 | HOXA5 | 11359 | -0.759 | -0.2218 | No | ||
| 47 | NXPH3 | 11416 | -0.772 | -0.2186 | No | ||
| 48 | TRIB1 | 11938 | -0.876 | -0.2398 | No | ||
| 49 | PERQ1 | 11994 | -0.885 | -0.2357 | No | ||
| 50 | TNXB | 12279 | -0.944 | -0.2435 | No | ||
| 51 | PRX | 12676 | -1.028 | -0.2566 | No | ||
| 52 | NRG2 | 13753 | -1.268 | -0.3046 | Yes | ||
| 53 | ARF6 | 13830 | -1.291 | -0.2984 | Yes | ||
| 54 | NRGN | 13912 | -1.308 | -0.2923 | Yes | ||
| 55 | HOXA9 | 14019 | -1.331 | -0.2874 | Yes | ||
| 56 | SERPINF2 | 14065 | -1.344 | -0.2791 | Yes | ||
| 57 | KIF1C | 14359 | -1.421 | -0.2836 | Yes | ||
| 58 | PKP3 | 14633 | -1.492 | -0.2864 | Yes | ||
| 59 | GGN | 14703 | -1.508 | -0.2781 | Yes | ||
| 60 | EPHB1 | 14878 | -1.556 | -0.2751 | Yes | ||
| 61 | MAP3K11 | 15378 | -1.708 | -0.2884 | Yes | ||
| 62 | SLITRK5 | 15450 | -1.731 | -0.2784 | Yes | ||
| 63 | YARS | 15736 | -1.831 | -0.2792 | Yes | ||
| 64 | PSCA | 15870 | -1.888 | -0.2713 | Yes | ||
| 65 | SCAMP3 | 16053 | -1.965 | -0.2655 | Yes | ||
| 66 | PGF | 16165 | -2.011 | -0.2554 | Yes | ||
| 67 | ATP5G2 | 16435 | -2.128 | -0.2530 | Yes | ||
| 68 | ADD3 | 16532 | -2.177 | -0.2408 | Yes | ||
| 69 | ERF | 16615 | -2.222 | -0.2275 | Yes | ||
| 70 | PPP1R9B | 16665 | -2.244 | -0.2122 | Yes | ||
| 71 | ASB7 | 16721 | -2.275 | -0.1971 | Yes | ||
| 72 | MNT | 16762 | -2.299 | -0.1809 | Yes | ||
| 73 | POFUT1 | 16969 | -2.428 | -0.1726 | Yes | ||
| 74 | EPN1 | 17395 | -2.732 | -0.1738 | Yes | ||
| 75 | DCTN1 | 17402 | -2.738 | -0.1522 | Yes | ||
| 76 | PRKAG1 | 17424 | -2.756 | -0.1314 | Yes | ||
| 77 | NEUROG1 | 17478 | -2.802 | -0.1119 | Yes | ||
| 78 | TITF1 | 17524 | -2.839 | -0.0917 | Yes | ||
| 79 | CTDSP1 | 17552 | -2.863 | -0.0703 | Yes | ||
| 80 | NKIRAS2 | 17893 | -3.246 | -0.0628 | Yes | ||
| 81 | ABL1 | 18257 | -3.934 | -0.0510 | Yes | ||
| 82 | ROM1 | 18302 | -4.073 | -0.0209 | Yes | ||
| 83 | PHKG2 | 18463 | -4.735 | 0.0083 | Yes |
