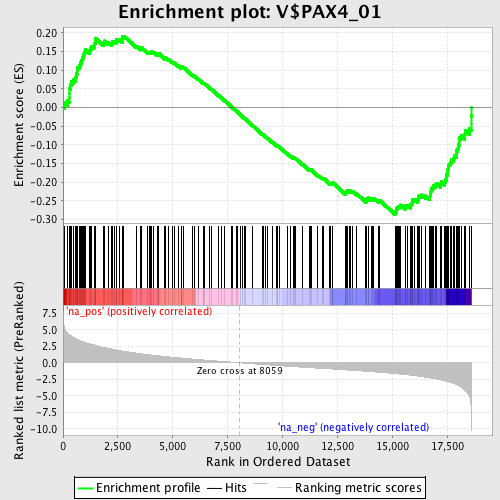
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$PAX4_01 |
| Enrichment Score (ES) | -0.28670025 |
| Normalized Enrichment Score (NES) | -1.3738604 |
| Nominal p-value | 0.016697587 |
| FDR q-value | 0.57255864 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC30A5 | 50 | 5.483 | 0.0133 | No | ||
| 2 | USP39 | 207 | 4.505 | 0.0179 | No | ||
| 3 | EED | 277 | 4.265 | 0.0266 | No | ||
| 4 | MXI1 | 280 | 4.258 | 0.0389 | No | ||
| 5 | TAF5 | 283 | 4.253 | 0.0512 | No | ||
| 6 | ELAVL1 | 334 | 4.139 | 0.0606 | No | ||
| 7 | KPNB1 | 374 | 4.033 | 0.0702 | No | ||
| 8 | TNRC15 | 471 | 3.835 | 0.0762 | No | ||
| 9 | ZDHHC5 | 574 | 3.656 | 0.0813 | No | ||
| 10 | MAML1 | 601 | 3.619 | 0.0904 | No | ||
| 11 | TPR | 635 | 3.575 | 0.0991 | No | ||
| 12 | RANBP1 | 654 | 3.537 | 0.1084 | No | ||
| 13 | EIF4A1 | 736 | 3.409 | 0.1139 | No | ||
| 14 | MAPRE1 | 785 | 3.327 | 0.1210 | No | ||
| 15 | GTF3C2 | 850 | 3.248 | 0.1270 | No | ||
| 16 | KLF13 | 885 | 3.203 | 0.1345 | No | ||
| 17 | ZCCHC9 | 932 | 3.150 | 0.1412 | No | ||
| 18 | APPBP1 | 967 | 3.110 | 0.1484 | No | ||
| 19 | MAPT | 1007 | 3.078 | 0.1553 | No | ||
| 20 | PABPC1 | 1215 | 2.865 | 0.1524 | No | ||
| 21 | ADIPOR2 | 1236 | 2.838 | 0.1596 | No | ||
| 22 | YWHAZ | 1292 | 2.796 | 0.1648 | No | ||
| 23 | MAPK10 | 1409 | 2.691 | 0.1663 | No | ||
| 24 | HIF3A | 1445 | 2.652 | 0.1721 | No | ||
| 25 | EIF2C2 | 1463 | 2.637 | 0.1789 | No | ||
| 26 | TAF11 | 1472 | 2.628 | 0.1861 | No | ||
| 27 | CTBP2 | 1847 | 2.322 | 0.1726 | No | ||
| 28 | SYNCRIP | 1864 | 2.313 | 0.1785 | No | ||
| 29 | PABPC4 | 2051 | 2.188 | 0.1748 | No | ||
| 30 | RBBP6 | 2208 | 2.077 | 0.1724 | No | ||
| 31 | WNT10A | 2245 | 2.056 | 0.1764 | No | ||
| 32 | GADD45G | 2321 | 1.999 | 0.1782 | No | ||
| 33 | PICALM | 2418 | 1.942 | 0.1786 | No | ||
| 34 | ADAMTSL1 | 2432 | 1.934 | 0.1835 | No | ||
| 35 | ANKMY2 | 2575 | 1.852 | 0.1812 | No | ||
| 36 | PSIP1 | 2689 | 1.781 | 0.1803 | No | ||
| 37 | NFATC4 | 2692 | 1.780 | 0.1854 | No | ||
| 38 | UBTF | 2702 | 1.772 | 0.1901 | No | ||
| 39 | DNAJC11 | 2770 | 1.732 | 0.1915 | No | ||
| 40 | E2F1 | 3349 | 1.457 | 0.1644 | No | ||
| 41 | H1FX | 3537 | 1.371 | 0.1582 | No | ||
| 42 | PDE4D | 3575 | 1.357 | 0.1602 | No | ||
| 43 | ACSL3 | 3861 | 1.230 | 0.1483 | No | ||
| 44 | NF2 | 3945 | 1.202 | 0.1473 | No | ||
| 45 | ISCU | 3985 | 1.185 | 0.1486 | No | ||
| 46 | NRD1 | 4012 | 1.174 | 0.1506 | No | ||
| 47 | CPNE1 | 4128 | 1.129 | 0.1477 | No | ||
| 48 | NR4A3 | 4300 | 1.070 | 0.1415 | No | ||
| 49 | ZFYVE27 | 4334 | 1.058 | 0.1428 | No | ||
| 50 | APBA2BP | 4351 | 1.049 | 0.1450 | No | ||
| 51 | IL17B | 4635 | 0.951 | 0.1324 | No | ||
| 52 | PLAGL2 | 4673 | 0.940 | 0.1332 | No | ||
| 53 | AMFR | 4785 | 0.901 | 0.1298 | No | ||
| 54 | ATOH1 | 4977 | 0.842 | 0.1219 | No | ||
| 55 | GTF2IRD1 | 5066 | 0.814 | 0.1195 | No | ||
| 56 | NFKBIA | 5250 | 0.761 | 0.1118 | No | ||
| 57 | EVX1 | 5377 | 0.718 | 0.1070 | No | ||
| 58 | COL27A1 | 5415 | 0.708 | 0.1071 | No | ||
| 59 | PCF11 | 5416 | 0.708 | 0.1091 | No | ||
| 60 | RBMS3 | 5475 | 0.686 | 0.1080 | No | ||
| 61 | SSH2 | 5898 | 0.569 | 0.0868 | No | ||
| 62 | HOXA7 | 5993 | 0.539 | 0.0832 | No | ||
| 63 | PHF12 | 6151 | 0.493 | 0.0762 | No | ||
| 64 | CDX1 | 6380 | 0.430 | 0.0650 | No | ||
| 65 | LIN28 | 6405 | 0.424 | 0.0650 | No | ||
| 66 | MMP11 | 6433 | 0.417 | 0.0647 | No | ||
| 67 | SPRY2 | 6663 | 0.354 | 0.0533 | No | ||
| 68 | PBX1 | 6757 | 0.333 | 0.0493 | No | ||
| 69 | LRP1 | 7097 | 0.238 | 0.0316 | No | ||
| 70 | XPO1 | 7218 | 0.210 | 0.0257 | No | ||
| 71 | SMAD1 | 7372 | 0.174 | 0.0179 | No | ||
| 72 | DNAJB5 | 7676 | 0.096 | 0.0017 | No | ||
| 73 | SLC4A1 | 7712 | 0.087 | 0.0001 | No | ||
| 74 | SLC25A28 | 7908 | 0.038 | -0.0104 | No | ||
| 75 | CPT1B | 7963 | 0.022 | -0.0133 | No | ||
| 76 | SP4 | 8066 | -0.002 | -0.0188 | No | ||
| 77 | PACS1 | 8183 | -0.029 | -0.0250 | No | ||
| 78 | ZIC2 | 8273 | -0.053 | -0.0297 | No | ||
| 79 | HMGN2 | 8310 | -0.061 | -0.0314 | No | ||
| 80 | RBP4 | 8642 | -0.147 | -0.0490 | No | ||
| 81 | KCNK13 | 9072 | -0.251 | -0.0715 | No | ||
| 82 | HOXD10 | 9116 | -0.262 | -0.0731 | No | ||
| 83 | PRDM1 | 9208 | -0.281 | -0.0772 | No | ||
| 84 | GALNT10 | 9314 | -0.303 | -0.0820 | No | ||
| 85 | GRIK1 | 9530 | -0.350 | -0.0927 | No | ||
| 86 | CCNI | 9711 | -0.393 | -0.1013 | No | ||
| 87 | JARID2 | 9757 | -0.404 | -0.1025 | No | ||
| 88 | EFNA5 | 9767 | -0.407 | -0.1018 | No | ||
| 89 | CDH16 | 9863 | -0.431 | -0.1057 | No | ||
| 90 | ZHX2 | 10220 | -0.512 | -0.1236 | No | ||
| 91 | IRX2 | 10377 | -0.547 | -0.1304 | No | ||
| 92 | CTF1 | 10493 | -0.574 | -0.1350 | No | ||
| 93 | SLC16A10 | 10529 | -0.582 | -0.1352 | No | ||
| 94 | PDGFB | 10575 | -0.591 | -0.1359 | No | ||
| 95 | TRIM3 | 10912 | -0.666 | -0.1522 | No | ||
| 96 | GRIN2A | 11231 | -0.732 | -0.1673 | No | ||
| 97 | ACOX2 | 11272 | -0.740 | -0.1673 | No | ||
| 98 | DDIT3 | 11304 | -0.746 | -0.1668 | No | ||
| 99 | HNRPR | 11614 | -0.811 | -0.1812 | No | ||
| 100 | MXD4 | 11805 | -0.849 | -0.1891 | No | ||
| 101 | PHF15 | 11875 | -0.863 | -0.1903 | No | ||
| 102 | C1QL1 | 12162 | -0.919 | -0.2031 | No | ||
| 103 | NRXN1 | 12208 | -0.928 | -0.2029 | No | ||
| 104 | TNXB | 12279 | -0.944 | -0.2039 | No | ||
| 105 | DLL4 | 12293 | -0.946 | -0.2019 | No | ||
| 106 | GNB2 | 12855 | -1.062 | -0.2292 | No | ||
| 107 | CACNA1D | 12869 | -1.064 | -0.2268 | No | ||
| 108 | SLC38A3 | 12909 | -1.073 | -0.2258 | No | ||
| 109 | PCTK1 | 12949 | -1.084 | -0.2248 | No | ||
| 110 | ARHGEF12 | 12966 | -1.086 | -0.2225 | No | ||
| 111 | NR0B2 | 13040 | -1.100 | -0.2232 | No | ||
| 112 | FGD1 | 13114 | -1.115 | -0.2239 | No | ||
| 113 | SEMA3B | 13200 | -1.135 | -0.2252 | No | ||
| 114 | GPR37L1 | 13373 | -1.174 | -0.2311 | No | ||
| 115 | RAB33A | 13798 | -1.282 | -0.2504 | No | ||
| 116 | CDX4 | 13813 | -1.286 | -0.2474 | No | ||
| 117 | ARF6 | 13830 | -1.291 | -0.2445 | No | ||
| 118 | BCL11A | 13924 | -1.312 | -0.2457 | No | ||
| 119 | BDNF | 13934 | -1.313 | -0.2424 | No | ||
| 120 | DNM1 | 14043 | -1.338 | -0.2443 | No | ||
| 121 | HOXA1 | 14116 | -1.357 | -0.2443 | No | ||
| 122 | SLC35B1 | 14168 | -1.369 | -0.2431 | No | ||
| 123 | CDK9 | 14383 | -1.427 | -0.2505 | No | ||
| 124 | BRUNOL6 | 14421 | -1.437 | -0.2483 | No | ||
| 125 | KLC2 | 15129 | -1.631 | -0.2819 | Yes | ||
| 126 | NTRK3 | 15169 | -1.644 | -0.2793 | Yes | ||
| 127 | RKHD3 | 15178 | -1.647 | -0.2749 | Yes | ||
| 128 | HOXB6 | 15192 | -1.651 | -0.2708 | Yes | ||
| 129 | PAQR4 | 15219 | -1.661 | -0.2674 | Yes | ||
| 130 | ESRRA | 15268 | -1.673 | -0.2651 | Yes | ||
| 131 | EFHD1 | 15347 | -1.700 | -0.2644 | Yes | ||
| 132 | AP1B1 | 15382 | -1.710 | -0.2612 | Yes | ||
| 133 | SORCS3 | 15585 | -1.771 | -0.2670 | Yes | ||
| 134 | ABHD2 | 15595 | -1.773 | -0.2623 | Yes | ||
| 135 | PER2 | 15701 | -1.815 | -0.2627 | Yes | ||
| 136 | CIC | 15841 | -1.875 | -0.2648 | Yes | ||
| 137 | SALL1 | 15849 | -1.878 | -0.2597 | Yes | ||
| 138 | TPM1 | 15894 | -1.894 | -0.2566 | Yes | ||
| 139 | PHKB | 15907 | -1.900 | -0.2517 | Yes | ||
| 140 | CIZ1 | 15924 | -1.906 | -0.2470 | Yes | ||
| 141 | DAAM2 | 16030 | -1.955 | -0.2470 | Yes | ||
| 142 | PGF | 16165 | -2.011 | -0.2484 | Yes | ||
| 143 | LEMD2 | 16188 | -2.022 | -0.2437 | Yes | ||
| 144 | CHKB | 16189 | -2.022 | -0.2378 | Yes | ||
| 145 | RRAS | 16241 | -2.041 | -0.2346 | Yes | ||
| 146 | STX1B2 | 16349 | -2.090 | -0.2344 | Yes | ||
| 147 | CNTFR | 16514 | -2.166 | -0.2369 | Yes | ||
| 148 | ANGPT1 | 16713 | -2.268 | -0.2411 | Yes | ||
| 149 | ASB7 | 16721 | -2.275 | -0.2348 | Yes | ||
| 150 | MNT | 16762 | -2.299 | -0.2303 | Yes | ||
| 151 | PTPRG | 16765 | -2.301 | -0.2237 | Yes | ||
| 152 | SART3 | 16780 | -2.315 | -0.2177 | Yes | ||
| 153 | HOXA10 | 16854 | -2.351 | -0.2148 | Yes | ||
| 154 | POU3F3 | 16887 | -2.372 | -0.2096 | Yes | ||
| 155 | POFUT1 | 16969 | -2.428 | -0.2070 | Yes | ||
| 156 | FBXO36 | 17031 | -2.468 | -0.2031 | Yes | ||
| 157 | TRERF1 | 17222 | -2.602 | -0.2058 | Yes | ||
| 158 | PIK3CD | 17239 | -2.615 | -0.1990 | Yes | ||
| 159 | BAT3 | 17404 | -2.739 | -0.1999 | Yes | ||
| 160 | PRKAG1 | 17424 | -2.756 | -0.1929 | Yes | ||
| 161 | LRP2 | 17456 | -2.779 | -0.1865 | Yes | ||
| 162 | NEUROG1 | 17478 | -2.802 | -0.1795 | Yes | ||
| 163 | TITF1 | 17524 | -2.839 | -0.1737 | Yes | ||
| 164 | AAMP | 17538 | -2.846 | -0.1661 | Yes | ||
| 165 | CTDSP1 | 17552 | -2.863 | -0.1584 | Yes | ||
| 166 | EIF5A | 17584 | -2.891 | -0.1517 | Yes | ||
| 167 | RING1 | 17660 | -2.983 | -0.1471 | Yes | ||
| 168 | PTPN23 | 17695 | -3.018 | -0.1401 | Yes | ||
| 169 | LBX1 | 17784 | -3.108 | -0.1358 | Yes | ||
| 170 | ETV5 | 17825 | -3.153 | -0.1288 | Yes | ||
| 171 | RPS6KA4 | 17922 | -3.288 | -0.1244 | Yes | ||
| 172 | HOXB1 | 17928 | -3.297 | -0.1151 | Yes | ||
| 173 | DGKZ | 17996 | -3.400 | -0.1088 | Yes | ||
| 174 | NCDN | 18013 | -3.437 | -0.0997 | Yes | ||
| 175 | PLAGL1 | 18046 | -3.494 | -0.0912 | Yes | ||
| 176 | SUOX | 18049 | -3.501 | -0.0811 | Yes | ||
| 177 | OAZ2 | 18134 | -3.654 | -0.0750 | Yes | ||
| 178 | FBXL19 | 18312 | -4.109 | -0.0726 | Yes | ||
| 179 | CAPN5 | 18321 | -4.145 | -0.0610 | Yes | ||
| 180 | HTF9C | 18515 | -5.047 | -0.0568 | Yes | ||
| 181 | RNF10 | 18596 | -6.596 | -0.0419 | Yes | ||
| 182 | HES1 | 18597 | -6.611 | -0.0226 | Yes | ||
| 183 | DTX1 | 18614 | -8.089 | 0.0001 | Yes |
