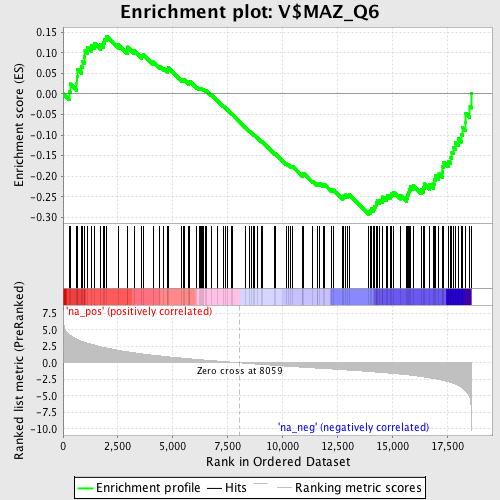
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MAZ_Q6 |
| Enrichment Score (ES) | -0.292708 |
| Normalized Enrichment Score (NES) | -1.3255451 |
| Nominal p-value | 0.040462427 |
| FDR q-value | 0.54386437 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SETDB1 | 272 | 4.276 | 0.0066 | No | ||
| 2 | CUGBP1 | 327 | 4.148 | 0.0243 | No | ||
| 3 | CNN3 | 630 | 3.581 | 0.0257 | No | ||
| 4 | GTF2H1 | 632 | 3.579 | 0.0435 | No | ||
| 5 | EIF4G2 | 660 | 3.525 | 0.0595 | No | ||
| 6 | TRIM8 | 847 | 3.256 | 0.0657 | No | ||
| 7 | TIAL1 | 894 | 3.194 | 0.0791 | No | ||
| 8 | SYT7 | 976 | 3.099 | 0.0901 | No | ||
| 9 | DDX50 | 996 | 3.082 | 0.1044 | No | ||
| 10 | ARID1A | 1116 | 2.949 | 0.1126 | No | ||
| 11 | TRIM33 | 1293 | 2.795 | 0.1170 | No | ||
| 12 | HSD11B1 | 1431 | 2.668 | 0.1229 | No | ||
| 13 | RPL5 | 1701 | 2.433 | 0.1204 | No | ||
| 14 | STX6 | 1835 | 2.336 | 0.1248 | No | ||
| 15 | HDAC1 | 1894 | 2.288 | 0.1331 | No | ||
| 16 | POU2F1 | 1967 | 2.246 | 0.1404 | No | ||
| 17 | MDM4 | 2519 | 1.882 | 0.1199 | No | ||
| 18 | UPF2 | 2929 | 1.648 | 0.1060 | No | ||
| 19 | MLLT10 | 2933 | 1.647 | 0.1140 | No | ||
| 20 | KCNIP2 | 3232 | 1.511 | 0.1054 | No | ||
| 21 | ATP1A1 | 3594 | 1.348 | 0.0926 | No | ||
| 22 | POGZ | 3644 | 1.319 | 0.0965 | No | ||
| 23 | KCNC1 | 4098 | 1.141 | 0.0777 | No | ||
| 24 | TCF7L2 | 4399 | 1.033 | 0.0666 | No | ||
| 25 | PRDM16 | 4576 | 0.971 | 0.0619 | No | ||
| 26 | RERE | 4773 | 0.905 | 0.0558 | No | ||
| 27 | MDK | 4776 | 0.904 | 0.0602 | No | ||
| 28 | TCEA3 | 4784 | 0.902 | 0.0643 | No | ||
| 29 | LHX4 | 5407 | 0.711 | 0.0342 | No | ||
| 30 | HNRPF | 5494 | 0.681 | 0.0329 | No | ||
| 31 | MGEA5 | 5536 | 0.669 | 0.0341 | No | ||
| 32 | ENSA | 5735 | 0.611 | 0.0264 | No | ||
| 33 | P4HA1 | 5755 | 0.606 | 0.0284 | No | ||
| 34 | HCRTR1 | 5761 | 0.603 | 0.0311 | No | ||
| 35 | MYOZ1 | 6091 | 0.509 | 0.0158 | No | ||
| 36 | JUN | 6199 | 0.481 | 0.0124 | No | ||
| 37 | KRTCAP2 | 6250 | 0.468 | 0.0121 | No | ||
| 38 | NEUROG3 | 6286 | 0.455 | 0.0124 | No | ||
| 39 | BRDT | 6339 | 0.443 | 0.0118 | No | ||
| 40 | LIN28 | 6405 | 0.424 | 0.0104 | No | ||
| 41 | EMX2 | 6487 | 0.403 | 0.0080 | No | ||
| 42 | NRAS | 6520 | 0.395 | 0.0083 | No | ||
| 43 | PBX1 | 6757 | 0.333 | -0.0028 | No | ||
| 44 | PIK3AP1 | 7035 | 0.254 | -0.0165 | No | ||
| 45 | NEURL | 7291 | 0.194 | -0.0294 | No | ||
| 46 | TLX1 | 7388 | 0.168 | -0.0337 | No | ||
| 47 | SYT11 | 7491 | 0.143 | -0.0385 | No | ||
| 48 | EFNA1 | 7677 | 0.096 | -0.0481 | No | ||
| 49 | PIP5K1A | 7726 | 0.085 | -0.0502 | No | ||
| 50 | HMGN2 | 8310 | -0.061 | -0.0815 | No | ||
| 51 | ASCL2 | 8514 | -0.118 | -0.0918 | No | ||
| 52 | XPR1 | 8597 | -0.139 | -0.0956 | No | ||
| 53 | PTPRJ | 8685 | -0.157 | -0.0995 | No | ||
| 54 | PPRC1 | 8733 | -0.169 | -0.1012 | No | ||
| 55 | KCNQ4 | 8837 | -0.196 | -0.1058 | No | ||
| 56 | ST5 | 9036 | -0.242 | -0.1153 | No | ||
| 57 | RNF2 | 9081 | -0.255 | -0.1164 | No | ||
| 58 | NFYC | 9650 | -0.379 | -0.1453 | No | ||
| 59 | F2 | 9659 | -0.382 | -0.1438 | No | ||
| 60 | CAMK2G | 10165 | -0.501 | -0.1686 | No | ||
| 61 | ROR1 | 10279 | -0.525 | -0.1721 | No | ||
| 62 | HPCA | 10384 | -0.550 | -0.1750 | No | ||
| 63 | RFX5 | 10442 | -0.562 | -0.1753 | No | ||
| 64 | SCUBE2 | 10907 | -0.665 | -0.1971 | No | ||
| 65 | PHF13 | 10920 | -0.667 | -0.1944 | No | ||
| 66 | PLCB3 | 10970 | -0.676 | -0.1937 | No | ||
| 67 | RTN4RL2 | 11377 | -0.764 | -0.2118 | No | ||
| 68 | CREM | 11594 | -0.807 | -0.2195 | No | ||
| 69 | HNRPR | 11614 | -0.811 | -0.2165 | No | ||
| 70 | THRAP3 | 11703 | -0.827 | -0.2172 | No | ||
| 71 | PLA2G2E | 11861 | -0.861 | -0.2214 | No | ||
| 72 | PROX1 | 11915 | -0.871 | -0.2199 | No | ||
| 73 | PAX6 | 12220 | -0.930 | -0.2317 | No | ||
| 74 | WAC | 12336 | -0.956 | -0.2332 | No | ||
| 75 | SLC2A1 | 12750 | -1.041 | -0.2503 | No | ||
| 76 | CUL2 | 12800 | -1.050 | -0.2478 | No | ||
| 77 | EPHA2 | 12852 | -1.062 | -0.2452 | No | ||
| 78 | BCL9 | 12974 | -1.087 | -0.2464 | No | ||
| 79 | NR0B2 | 13040 | -1.100 | -0.2444 | No | ||
| 80 | BDNF | 13934 | -1.313 | -0.2862 | Yes | ||
| 81 | TRIM46 | 14017 | -1.331 | -0.2840 | Yes | ||
| 82 | SSBP3 | 14046 | -1.339 | -0.2789 | Yes | ||
| 83 | APBB1 | 14164 | -1.369 | -0.2784 | Yes | ||
| 84 | FDPS | 14190 | -1.375 | -0.2729 | Yes | ||
| 85 | GALNT2 | 14260 | -1.393 | -0.2697 | Yes | ||
| 86 | PRELP | 14277 | -1.398 | -0.2636 | Yes | ||
| 87 | MYBPC3 | 14322 | -1.410 | -0.2590 | Yes | ||
| 88 | JMJD2A | 14439 | -1.440 | -0.2581 | Yes | ||
| 89 | GRK5 | 14544 | -1.469 | -0.2564 | Yes | ||
| 90 | PACSIN3 | 14564 | -1.473 | -0.2501 | Yes | ||
| 91 | SHC1 | 14730 | -1.514 | -0.2515 | Yes | ||
| 92 | PTPN5 | 14792 | -1.530 | -0.2472 | Yes | ||
| 93 | DUSP8 | 14906 | -1.566 | -0.2455 | Yes | ||
| 94 | FKBP2 | 14987 | -1.588 | -0.2419 | Yes | ||
| 95 | SESN2 | 15066 | -1.611 | -0.2382 | Yes | ||
| 96 | ARHGEF2 | 15383 | -1.710 | -0.2467 | Yes | ||
| 97 | TNNI2 | 15652 | -1.792 | -0.2523 | Yes | ||
| 98 | NPPA | 15684 | -1.807 | -0.2450 | Yes | ||
| 99 | YARS | 15736 | -1.831 | -0.2387 | Yes | ||
| 100 | LCK | 15768 | -1.842 | -0.2312 | Yes | ||
| 101 | GRIK3 | 15823 | -1.867 | -0.2248 | Yes | ||
| 102 | NR1H3 | 15951 | -1.919 | -0.2221 | Yes | ||
| 103 | CLSTN1 | 16327 | -2.080 | -0.2321 | Yes | ||
| 104 | FHL3 | 16443 | -2.131 | -0.2277 | Yes | ||
| 105 | EPHB2 | 16469 | -2.145 | -0.2184 | Yes | ||
| 106 | PLK3 | 16716 | -2.270 | -0.2204 | Yes | ||
| 107 | FXC1 | 16897 | -2.378 | -0.2183 | Yes | ||
| 108 | CASQ1 | 16933 | -2.402 | -0.2083 | Yes | ||
| 109 | UBE4B | 16971 | -2.430 | -0.1982 | Yes | ||
| 110 | BAI2 | 17131 | -2.534 | -0.1942 | Yes | ||
| 111 | PIAS3 | 17287 | -2.648 | -0.1894 | Yes | ||
| 112 | ARFIP2 | 17305 | -2.666 | -0.1771 | Yes | ||
| 113 | OTUB1 | 17337 | -2.694 | -0.1653 | Yes | ||
| 114 | DNAJB12 | 17560 | -2.870 | -0.1631 | Yes | ||
| 115 | PPOX | 17675 | -2.998 | -0.1543 | Yes | ||
| 116 | TPM3 | 17718 | -3.040 | -0.1415 | Yes | ||
| 117 | LBX1 | 17784 | -3.108 | -0.1295 | Yes | ||
| 118 | GFRA1 | 17875 | -3.221 | -0.1184 | Yes | ||
| 119 | NCDN | 18013 | -3.437 | -0.1087 | Yes | ||
| 120 | PHC2 | 18165 | -3.704 | -0.0985 | Yes | ||
| 121 | LMNA | 18217 | -3.800 | -0.0823 | Yes | ||
| 122 | PTPRF | 18349 | -4.251 | -0.0683 | Yes | ||
| 123 | LRRC20 | 18356 | -4.275 | -0.0473 | Yes | ||
| 124 | RHOG | 18544 | -5.301 | -0.0311 | Yes | ||
| 125 | MOV10 | 18603 | -7.025 | 0.0007 | Yes |
