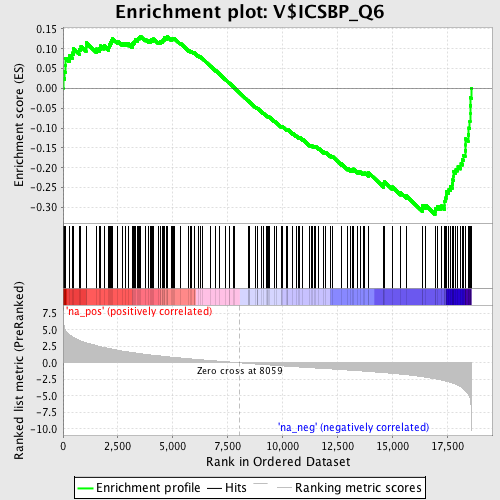
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ICSBP_Q6 |
| Enrichment Score (ES) | -0.31780896 |
| Normalized Enrichment Score (NES) | -1.5006807 |
| Nominal p-value | 0.0037105752 |
| FDR q-value | 0.4420823 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 5 | 7.263 | 0.0262 | No | ||
| 2 | ITPR1 | 69 | 5.257 | 0.0420 | No | ||
| 3 | PSMA3 | 97 | 4.977 | 0.0587 | No | ||
| 4 | EMP1 | 107 | 4.902 | 0.0761 | No | ||
| 5 | MXI1 | 280 | 4.258 | 0.0824 | No | ||
| 6 | PIK3R3 | 416 | 3.932 | 0.0894 | No | ||
| 7 | CXCR4 | 479 | 3.818 | 0.1000 | No | ||
| 8 | CHST1 | 745 | 3.397 | 0.0980 | No | ||
| 9 | LY86 | 812 | 3.295 | 0.1065 | No | ||
| 10 | USP18 | 1048 | 3.035 | 0.1049 | No | ||
| 11 | RRBP1 | 1054 | 3.025 | 0.1156 | No | ||
| 12 | LGALS8 | 1526 | 2.575 | 0.0995 | No | ||
| 13 | TOP1 | 1664 | 2.462 | 0.1011 | No | ||
| 14 | BBX | 1714 | 2.426 | 0.1073 | No | ||
| 15 | SYNCRIP | 1864 | 2.313 | 0.1077 | No | ||
| 16 | PHF6 | 2088 | 2.166 | 0.1035 | No | ||
| 17 | NT5C3 | 2098 | 2.160 | 0.1109 | No | ||
| 18 | CTSS | 2150 | 2.116 | 0.1158 | No | ||
| 19 | RIPK2 | 2192 | 2.088 | 0.1212 | No | ||
| 20 | IL18BP | 2259 | 2.048 | 0.1251 | No | ||
| 21 | LRMP | 2496 | 1.897 | 0.1193 | No | ||
| 22 | PKP4 | 2708 | 1.767 | 0.1143 | No | ||
| 23 | CYP7A1 | 2823 | 1.706 | 0.1144 | No | ||
| 24 | FES | 2962 | 1.634 | 0.1129 | No | ||
| 25 | MAP2K6 | 3157 | 1.542 | 0.1080 | No | ||
| 26 | ARHGEF6 | 3172 | 1.535 | 0.1128 | No | ||
| 27 | DAPP1 | 3218 | 1.516 | 0.1159 | No | ||
| 28 | SIX1 | 3270 | 1.496 | 0.1186 | No | ||
| 29 | ATP10A | 3286 | 1.486 | 0.1232 | No | ||
| 30 | VGLL4 | 3381 | 1.444 | 0.1234 | No | ||
| 31 | HNRPD | 3413 | 1.430 | 0.1269 | No | ||
| 32 | MGAT4A | 3460 | 1.403 | 0.1296 | No | ||
| 33 | CASK | 3512 | 1.379 | 0.1319 | No | ||
| 34 | THRAP1 | 3741 | 1.280 | 0.1242 | No | ||
| 35 | ATP5H | 3887 | 1.220 | 0.1208 | No | ||
| 36 | FUT8 | 3994 | 1.181 | 0.1193 | No | ||
| 37 | PPARGC1A | 4010 | 1.174 | 0.1228 | No | ||
| 38 | ATP11C | 4089 | 1.145 | 0.1228 | No | ||
| 39 | AIF1 | 4126 | 1.130 | 0.1249 | No | ||
| 40 | ZBP1 | 4345 | 1.052 | 0.1170 | No | ||
| 41 | LY75 | 4436 | 1.022 | 0.1158 | No | ||
| 42 | GATA6 | 4448 | 1.018 | 0.1190 | No | ||
| 43 | NDRG3 | 4525 | 0.989 | 0.1184 | No | ||
| 44 | MPDZ | 4545 | 0.983 | 0.1210 | No | ||
| 45 | PRDM16 | 4576 | 0.971 | 0.1229 | No | ||
| 46 | ELK4 | 4606 | 0.961 | 0.1249 | No | ||
| 47 | TLR7 | 4614 | 0.959 | 0.1280 | No | ||
| 48 | KYNU | 4710 | 0.928 | 0.1262 | No | ||
| 49 | LAMA3 | 4741 | 0.917 | 0.1279 | No | ||
| 50 | HIST1H1C | 4751 | 0.913 | 0.1308 | No | ||
| 51 | TFEC | 4951 | 0.852 | 0.1231 | No | ||
| 52 | IL22RA1 | 4972 | 0.843 | 0.1251 | No | ||
| 53 | ZBTB33 | 5010 | 0.828 | 0.1261 | No | ||
| 54 | NPAS3 | 5063 | 0.815 | 0.1263 | No | ||
| 55 | LRRN3 | 5333 | 0.733 | 0.1144 | No | ||
| 56 | CSAD | 5726 | 0.614 | 0.0954 | No | ||
| 57 | BST2 | 5800 | 0.595 | 0.0936 | No | ||
| 58 | ESR1 | 5859 | 0.578 | 0.0926 | No | ||
| 59 | DGKA | 5967 | 0.546 | 0.0888 | No | ||
| 60 | ENPP2 | 5975 | 0.544 | 0.0904 | No | ||
| 61 | CDK6 | 6164 | 0.489 | 0.0820 | No | ||
| 62 | CXCL16 | 6241 | 0.469 | 0.0796 | No | ||
| 63 | TNFRSF17 | 6338 | 0.443 | 0.0760 | No | ||
| 64 | USP28 | 6725 | 0.341 | 0.0563 | No | ||
| 65 | SLITRK6 | 6962 | 0.276 | 0.0446 | No | ||
| 66 | IL21 | 7123 | 0.232 | 0.0367 | No | ||
| 67 | DIO2 | 7383 | 0.170 | 0.0233 | No | ||
| 68 | TNFSF15 | 7581 | 0.121 | 0.0131 | No | ||
| 69 | OSR2 | 7591 | 0.118 | 0.0130 | No | ||
| 70 | NPR3 | 7759 | 0.077 | 0.0043 | No | ||
| 71 | HAPLN1 | 7831 | 0.061 | 0.0006 | No | ||
| 72 | OTX1 | 8430 | -0.095 | -0.0314 | No | ||
| 73 | TCF15 | 8461 | -0.106 | -0.0326 | No | ||
| 74 | RCOR1 | 8494 | -0.114 | -0.0340 | No | ||
| 75 | ASCL2 | 8514 | -0.118 | -0.0346 | No | ||
| 76 | ALDH1A1 | 8771 | -0.180 | -0.0478 | No | ||
| 77 | GPR22 | 8841 | -0.197 | -0.0508 | No | ||
| 78 | HAS3 | 8853 | -0.200 | -0.0507 | No | ||
| 79 | IFNB1 | 8864 | -0.202 | -0.0505 | No | ||
| 80 | BANK1 | 9042 | -0.245 | -0.0592 | No | ||
| 81 | HOXA13 | 9119 | -0.262 | -0.0623 | No | ||
| 82 | CBX4 | 9252 | -0.289 | -0.0684 | No | ||
| 83 | NKX2-2 | 9323 | -0.304 | -0.0711 | No | ||
| 84 | INDO | 9335 | -0.306 | -0.0706 | No | ||
| 85 | SOCS1 | 9375 | -0.314 | -0.0715 | No | ||
| 86 | SATB2 | 9416 | -0.321 | -0.0725 | No | ||
| 87 | TEK | 9630 | -0.373 | -0.0827 | No | ||
| 88 | IRAK1 | 9710 | -0.393 | -0.0856 | No | ||
| 89 | PURA | 9948 | -0.451 | -0.0967 | No | ||
| 90 | PAX3 | 9951 | -0.451 | -0.0952 | No | ||
| 91 | KLHL13 | 9980 | -0.459 | -0.0950 | No | ||
| 92 | SEMA6D | 10187 | -0.506 | -0.1044 | No | ||
| 93 | LGI1 | 10228 | -0.513 | -0.1047 | No | ||
| 94 | TNFSF13B | 10238 | -0.514 | -0.1033 | No | ||
| 95 | ASPA | 10473 | -0.570 | -0.1139 | No | ||
| 96 | IL27 | 10624 | -0.601 | -0.1198 | No | ||
| 97 | SLC15A3 | 10738 | -0.627 | -0.1236 | No | ||
| 98 | HOXA3 | 10787 | -0.638 | -0.1239 | No | ||
| 99 | ERBB2 | 10915 | -0.666 | -0.1283 | No | ||
| 100 | ADAM8 | 11232 | -0.732 | -0.1428 | No | ||
| 101 | FGF9 | 11311 | -0.747 | -0.1443 | No | ||
| 102 | ZIC5 | 11376 | -0.764 | -0.1450 | No | ||
| 103 | NFKB1 | 11453 | -0.779 | -0.1462 | No | ||
| 104 | ZFPM1 | 11499 | -0.786 | -0.1458 | No | ||
| 105 | SOS1 | 11653 | -0.819 | -0.1511 | No | ||
| 106 | PHF15 | 11875 | -0.863 | -0.1599 | No | ||
| 107 | SLC11A2 | 11965 | -0.880 | -0.1615 | No | ||
| 108 | PPP2R5C | 12176 | -0.921 | -0.1695 | No | ||
| 109 | HIVEP1 | 12283 | -0.945 | -0.1718 | No | ||
| 110 | COL4A1 | 12686 | -1.030 | -0.1899 | No | ||
| 111 | POU4F1 | 12980 | -1.089 | -0.2018 | No | ||
| 112 | COL4A2 | 13112 | -1.114 | -0.2048 | No | ||
| 113 | MSX1 | 13198 | -1.135 | -0.2052 | No | ||
| 114 | FMR1 | 13235 | -1.144 | -0.2030 | No | ||
| 115 | GZMK | 13430 | -1.191 | -0.2092 | No | ||
| 116 | EREG | 13537 | -1.216 | -0.2105 | No | ||
| 117 | TNRC4 | 13673 | -1.250 | -0.2132 | No | ||
| 118 | CASP7 | 13756 | -1.268 | -0.2130 | No | ||
| 119 | TRPM3 | 13921 | -1.311 | -0.2171 | No | ||
| 120 | BCL11A | 13924 | -1.312 | -0.2125 | No | ||
| 121 | ELAVL2 | 14602 | -1.483 | -0.2437 | No | ||
| 122 | TBX1 | 14610 | -1.486 | -0.2387 | No | ||
| 123 | ENTPD1 | 14631 | -1.492 | -0.2343 | No | ||
| 124 | THBS1 | 14996 | -1.590 | -0.2482 | No | ||
| 125 | MAP3K11 | 15378 | -1.708 | -0.2626 | No | ||
| 126 | PRDX5 | 15645 | -1.791 | -0.2705 | No | ||
| 127 | ECM2 | 16372 | -2.101 | -0.3022 | No | ||
| 128 | KCNJ1 | 16388 | -2.112 | -0.2953 | No | ||
| 129 | CNTFR | 16514 | -2.166 | -0.2941 | No | ||
| 130 | SP8 | 16952 | -2.417 | -0.3090 | Yes | ||
| 131 | HOXB7 | 16981 | -2.438 | -0.3016 | Yes | ||
| 132 | CDKN2C | 17060 | -2.484 | -0.2968 | Yes | ||
| 133 | TWIST1 | 17225 | -2.604 | -0.2961 | Yes | ||
| 134 | EPN1 | 17395 | -2.732 | -0.2953 | Yes | ||
| 135 | ITPR3 | 17398 | -2.734 | -0.2854 | Yes | ||
| 136 | PIGR | 17412 | -2.747 | -0.2761 | Yes | ||
| 137 | LRP2 | 17456 | -2.779 | -0.2683 | Yes | ||
| 138 | ADAR | 17477 | -2.801 | -0.2592 | Yes | ||
| 139 | EIF5A | 17584 | -2.891 | -0.2543 | Yes | ||
| 140 | USF1 | 17659 | -2.983 | -0.2475 | Yes | ||
| 141 | CABLES1 | 17732 | -3.054 | -0.2402 | Yes | ||
| 142 | ANGPTL4 | 17763 | -3.090 | -0.2306 | Yes | ||
| 143 | PSMB8 | 17796 | -3.119 | -0.2209 | Yes | ||
| 144 | PSMB9 | 17804 | -3.136 | -0.2098 | Yes | ||
| 145 | HPCAL1 | 17904 | -3.260 | -0.2033 | Yes | ||
| 146 | TNFRSF19 | 17997 | -3.401 | -0.1959 | Yes | ||
| 147 | DUSP10 | 18133 | -3.652 | -0.1898 | Yes | ||
| 148 | TAP1 | 18196 | -3.763 | -0.1795 | Yes | ||
| 149 | SLC12A7 | 18251 | -3.922 | -0.1681 | Yes | ||
| 150 | UBE1L | 18322 | -4.147 | -0.1567 | Yes | ||
| 151 | GSDMDC1 | 18346 | -4.236 | -0.1425 | Yes | ||
| 152 | TCIRG1 | 18351 | -4.259 | -0.1272 | Yes | ||
| 153 | STAT6 | 18459 | -4.707 | -0.1158 | Yes | ||
| 154 | B2M | 18496 | -4.920 | -0.0998 | Yes | ||
| 155 | TAPBP | 18539 | -5.241 | -0.0829 | Yes | ||
| 156 | SP110 | 18549 | -5.440 | -0.0635 | Yes | ||
| 157 | IFI35 | 18555 | -5.547 | -0.0435 | Yes | ||
| 158 | DDB2 | 18574 | -5.775 | -0.0234 | Yes | ||
| 159 | MOV10 | 18603 | -7.025 | 0.0007 | Yes |
