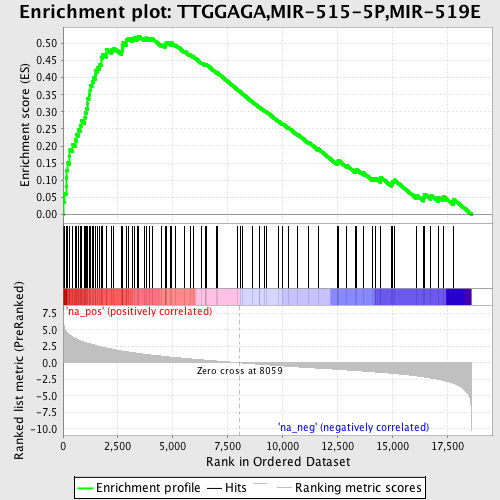
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | 0.5215831 |
| Normalized Enrichment Score (NES) | 2.3064432 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FREQ | 8 | 6.962 | 0.0363 | Yes | ||
| 2 | ICK | 66 | 5.291 | 0.0612 | Yes | ||
| 3 | AP3S1 | 137 | 4.726 | 0.0823 | Yes | ||
| 4 | RPP14 | 146 | 4.699 | 0.1067 | Yes | ||
| 5 | TMEM2 | 173 | 4.619 | 0.1297 | Yes | ||
| 6 | SFRS1 | 209 | 4.486 | 0.1515 | Yes | ||
| 7 | RBL2 | 302 | 4.213 | 0.1688 | Yes | ||
| 8 | ZFP36L1 | 311 | 4.187 | 0.1904 | Yes | ||
| 9 | UHRF2 | 441 | 3.890 | 0.2040 | Yes | ||
| 10 | RAP1B | 546 | 3.696 | 0.2179 | Yes | ||
| 11 | RIOK3 | 623 | 3.591 | 0.2328 | Yes | ||
| 12 | YAF2 | 697 | 3.470 | 0.2471 | Yes | ||
| 13 | MAPRE1 | 785 | 3.327 | 0.2600 | Yes | ||
| 14 | DDX3X | 840 | 3.265 | 0.2743 | Yes | ||
| 15 | CMTM6 | 994 | 3.085 | 0.2823 | Yes | ||
| 16 | RAP2C | 1021 | 3.061 | 0.2971 | Yes | ||
| 17 | EPC2 | 1078 | 3.007 | 0.3099 | Yes | ||
| 18 | ARID1A | 1116 | 2.949 | 0.3235 | Yes | ||
| 19 | UBAP2 | 1129 | 2.938 | 0.3384 | Yes | ||
| 20 | TNPO1 | 1180 | 2.895 | 0.3510 | Yes | ||
| 21 | SUMO2 | 1224 | 2.853 | 0.3637 | Yes | ||
| 22 | YTHDF2 | 1234 | 2.842 | 0.3782 | Yes | ||
| 23 | ZBTB1 | 1321 | 2.771 | 0.3882 | Yes | ||
| 24 | PUM1 | 1370 | 2.724 | 0.4000 | Yes | ||
| 25 | RDH11 | 1474 | 2.625 | 0.4083 | Yes | ||
| 26 | MARCKS | 1488 | 2.603 | 0.4213 | Yes | ||
| 27 | UBE2H | 1584 | 2.527 | 0.4295 | Yes | ||
| 28 | SLC7A11 | 1671 | 2.458 | 0.4379 | Yes | ||
| 29 | NCOA1 | 1735 | 2.409 | 0.4472 | Yes | ||
| 30 | HNRPUL1 | 1736 | 2.408 | 0.4599 | Yes | ||
| 31 | HIP2 | 1808 | 2.357 | 0.4685 | Yes | ||
| 32 | KPNA1 | 1965 | 2.248 | 0.4719 | Yes | ||
| 33 | POU2F1 | 1967 | 2.246 | 0.4838 | Yes | ||
| 34 | MTHFD1L | 2219 | 2.071 | 0.4811 | Yes | ||
| 35 | CYR61 | 2314 | 2.003 | 0.4866 | Yes | ||
| 36 | EPN2 | 2657 | 1.803 | 0.4777 | Yes | ||
| 37 | NFATC4 | 2692 | 1.780 | 0.4852 | Yes | ||
| 38 | SOX12 | 2704 | 1.770 | 0.4940 | Yes | ||
| 39 | IQGAP1 | 2728 | 1.760 | 0.5020 | Yes | ||
| 40 | PDGFRA | 2899 | 1.665 | 0.5016 | Yes | ||
| 41 | ATXN3 | 2901 | 1.664 | 0.5104 | Yes | ||
| 42 | BTG2 | 2959 | 1.634 | 0.5159 | Yes | ||
| 43 | ULK2 | 3146 | 1.548 | 0.5141 | Yes | ||
| 44 | SGPL1 | 3233 | 1.511 | 0.5174 | Yes | ||
| 45 | ETV1 | 3371 | 1.448 | 0.5176 | Yes | ||
| 46 | RPS6KA3 | 3437 | 1.415 | 0.5216 | Yes | ||
| 47 | FRAS1 | 3697 | 1.299 | 0.5145 | No | ||
| 48 | ATRN | 3786 | 1.265 | 0.5164 | No | ||
| 49 | DONSON | 3941 | 1.204 | 0.5144 | No | ||
| 50 | ABCG4 | 4070 | 1.156 | 0.5136 | No | ||
| 51 | RSBN1 | 4477 | 1.009 | 0.4970 | No | ||
| 52 | OPHN1 | 4646 | 0.948 | 0.4929 | No | ||
| 53 | PLAGL2 | 4673 | 0.940 | 0.4965 | No | ||
| 54 | ARHGEF3 | 4680 | 0.938 | 0.5011 | No | ||
| 55 | NOTCH2 | 4727 | 0.921 | 0.5035 | No | ||
| 56 | DDX3Y | 4895 | 0.868 | 0.4991 | No | ||
| 57 | OSBPL3 | 4924 | 0.860 | 0.5021 | No | ||
| 58 | TMEM47 | 5129 | 0.795 | 0.4953 | No | ||
| 59 | SYNGR1 | 5543 | 0.666 | 0.4765 | No | ||
| 60 | MOSPD2 | 5786 | 0.599 | 0.4666 | No | ||
| 61 | ARRDC3 | 5940 | 0.555 | 0.4612 | No | ||
| 62 | ISL1 | 6328 | 0.444 | 0.4427 | No | ||
| 63 | EMX2 | 6487 | 0.403 | 0.4363 | No | ||
| 64 | BSN | 6488 | 0.403 | 0.4384 | No | ||
| 65 | SNAP25 | 6512 | 0.397 | 0.4392 | No | ||
| 66 | UBE2B | 7000 | 0.263 | 0.4143 | No | ||
| 67 | ANUBL1 | 7019 | 0.259 | 0.4147 | No | ||
| 68 | ETNK2 | 7951 | 0.026 | 0.3646 | No | ||
| 69 | SP4 | 8066 | -0.002 | 0.3584 | No | ||
| 70 | MTF2 | 8155 | -0.021 | 0.3538 | No | ||
| 71 | CAB39 | 8650 | -0.150 | 0.3279 | No | ||
| 72 | ABCC3 | 8946 | -0.219 | 0.3131 | No | ||
| 73 | ACY1L2 | 8952 | -0.219 | 0.3140 | No | ||
| 74 | ITSN1 | 9186 | -0.277 | 0.3029 | No | ||
| 75 | KCTD7 | 9258 | -0.290 | 0.3006 | No | ||
| 76 | MEF2D | 9812 | -0.420 | 0.2729 | No | ||
| 77 | NFIB | 9981 | -0.460 | 0.2663 | No | ||
| 78 | CLCF1 | 10253 | -0.519 | 0.2544 | No | ||
| 79 | CPEB2 | 10697 | -0.619 | 0.2337 | No | ||
| 80 | FSTL1 | 11177 | -0.721 | 0.2117 | No | ||
| 81 | SOX30 | 11618 | -0.811 | 0.1922 | No | ||
| 82 | APLN | 12489 | -0.987 | 0.1504 | No | ||
| 83 | SLC9A2 | 12519 | -0.993 | 0.1541 | No | ||
| 84 | SLC4A4 | 12532 | -0.996 | 0.1587 | No | ||
| 85 | GDI1 | 12922 | -1.077 | 0.1434 | No | ||
| 86 | MLLT6 | 13333 | -1.164 | 0.1274 | No | ||
| 87 | POM121 | 13359 | -1.171 | 0.1322 | No | ||
| 88 | KCND2 | 13678 | -1.252 | 0.1217 | No | ||
| 89 | HOXA1 | 14116 | -1.357 | 0.1052 | No | ||
| 90 | FGFR2 | 14249 | -1.390 | 0.1054 | No | ||
| 91 | DCX | 14465 | -1.447 | 0.1014 | No | ||
| 92 | ETS1 | 14475 | -1.449 | 0.1086 | No | ||
| 93 | SERTAD1 | 14961 | -1.580 | 0.0908 | No | ||
| 94 | NUP35 | 15020 | -1.597 | 0.0961 | No | ||
| 95 | SCN8A | 15095 | -1.622 | 0.1006 | No | ||
| 96 | CSNK1G1 | 16111 | -1.987 | 0.0563 | No | ||
| 97 | CLSTN2 | 16434 | -2.128 | 0.0501 | No | ||
| 98 | PSMD11 | 16480 | -2.148 | 0.0590 | No | ||
| 99 | GABBR1 | 16763 | -2.299 | 0.0559 | No | ||
| 100 | TGFBR2 | 17108 | -2.516 | 0.0507 | No | ||
| 101 | EFNA3 | 17319 | -2.680 | 0.0535 | No | ||
| 102 | DGCR2 | 17803 | -3.133 | 0.0439 | No |