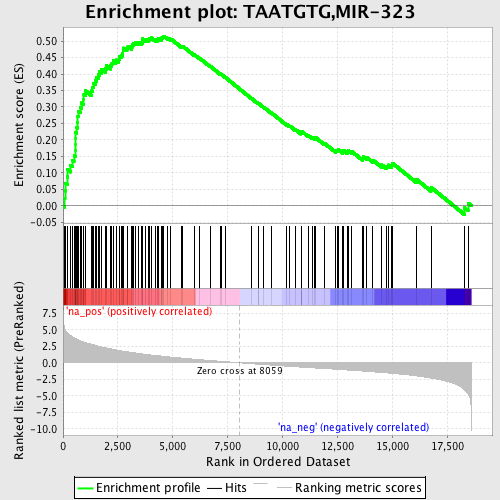
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | 0.5150608 |
| Normalized Enrichment Score (NES) | 2.2920027 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SATB1 | 70 | 5.244 | 0.0225 | Yes | ||
| 2 | HS2ST1 | 87 | 5.048 | 0.0469 | Yes | ||
| 3 | TMPO | 124 | 4.809 | 0.0690 | Yes | ||
| 4 | NDEL1 | 194 | 4.555 | 0.0880 | Yes | ||
| 5 | SFRS1 | 209 | 4.486 | 0.1097 | Yes | ||
| 6 | SUMO1 | 342 | 4.114 | 0.1232 | Yes | ||
| 7 | UHRF2 | 441 | 3.890 | 0.1374 | Yes | ||
| 8 | UBL3 | 499 | 3.780 | 0.1532 | Yes | ||
| 9 | RAB1A | 560 | 3.680 | 0.1684 | Yes | ||
| 10 | ARFIP1 | 564 | 3.677 | 0.1866 | Yes | ||
| 11 | PITPNA | 567 | 3.673 | 0.2049 | Yes | ||
| 12 | RANBP9 | 570 | 3.662 | 0.2231 | Yes | ||
| 13 | ZNF318 | 624 | 3.591 | 0.2382 | Yes | ||
| 14 | PPP1CB | 666 | 3.512 | 0.2536 | Yes | ||
| 15 | SLMAP | 674 | 3.502 | 0.2707 | Yes | ||
| 16 | HIPK1 | 686 | 3.486 | 0.2876 | Yes | ||
| 17 | MAPRE1 | 785 | 3.327 | 0.2989 | Yes | ||
| 18 | DDX3X | 840 | 3.265 | 0.3123 | Yes | ||
| 19 | CREB1 | 943 | 3.133 | 0.3225 | Yes | ||
| 20 | REV3L | 948 | 3.128 | 0.3379 | Yes | ||
| 21 | NFIX | 1025 | 3.058 | 0.3491 | Yes | ||
| 22 | TRIM33 | 1293 | 2.795 | 0.3487 | Yes | ||
| 23 | CLASP1 | 1347 | 2.744 | 0.3596 | Yes | ||
| 24 | TNRC6A | 1374 | 2.721 | 0.3718 | Yes | ||
| 25 | DHX36 | 1466 | 2.636 | 0.3801 | Yes | ||
| 26 | GJA1 | 1527 | 2.575 | 0.3897 | Yes | ||
| 27 | USP9X | 1604 | 2.508 | 0.3981 | Yes | ||
| 28 | TOP1 | 1664 | 2.462 | 0.4073 | Yes | ||
| 29 | SMARCAD1 | 1756 | 2.394 | 0.4143 | Yes | ||
| 30 | KPNA3 | 1922 | 2.272 | 0.4168 | Yes | ||
| 31 | KPNA1 | 1965 | 2.248 | 0.4258 | Yes | ||
| 32 | CLCN3 | 2164 | 2.107 | 0.4256 | Yes | ||
| 33 | ZHX1 | 2225 | 2.067 | 0.4327 | Yes | ||
| 34 | UTX | 2273 | 2.031 | 0.4403 | Yes | ||
| 35 | CEPT1 | 2410 | 1.949 | 0.4427 | Yes | ||
| 36 | MRC1 | 2547 | 1.866 | 0.4447 | Yes | ||
| 37 | FEM1B | 2563 | 1.857 | 0.4532 | Yes | ||
| 38 | SLITRK2 | 2648 | 1.808 | 0.4577 | Yes | ||
| 39 | PKP4 | 2708 | 1.767 | 0.4634 | Yes | ||
| 40 | CUL1 | 2729 | 1.757 | 0.4711 | Yes | ||
| 41 | RAB10 | 2745 | 1.750 | 0.4790 | Yes | ||
| 42 | TOP2B | 2913 | 1.659 | 0.4783 | Yes | ||
| 43 | CSNK1G3 | 2955 | 1.636 | 0.4843 | Yes | ||
| 44 | ZDHHC21 | 3131 | 1.553 | 0.4826 | Yes | ||
| 45 | ULK2 | 3146 | 1.548 | 0.4896 | Yes | ||
| 46 | TBPL1 | 3207 | 1.519 | 0.4940 | Yes | ||
| 47 | MAP4K4 | 3316 | 1.473 | 0.4955 | Yes | ||
| 48 | RPS6KA3 | 3437 | 1.415 | 0.4961 | Yes | ||
| 49 | PJA2 | 3570 | 1.359 | 0.4958 | Yes | ||
| 50 | PCDH9 | 3616 | 1.333 | 0.5000 | Yes | ||
| 51 | ASF1A | 3617 | 1.333 | 0.5067 | Yes | ||
| 52 | FGD4 | 3768 | 1.270 | 0.5049 | Yes | ||
| 53 | WBP2 | 3897 | 1.217 | 0.5041 | Yes | ||
| 54 | HOOK3 | 3922 | 1.211 | 0.5089 | Yes | ||
| 55 | PPARGC1A | 4010 | 1.174 | 0.5100 | Yes | ||
| 56 | CCND1 | 4211 | 1.103 | 0.5047 | Yes | ||
| 57 | NR4A3 | 4300 | 1.070 | 0.5053 | Yes | ||
| 58 | PDCD10 | 4342 | 1.054 | 0.5084 | Yes | ||
| 59 | RSBN1 | 4477 | 1.009 | 0.5062 | Yes | ||
| 60 | DCUN1D1 | 4504 | 0.998 | 0.5098 | Yes | ||
| 61 | CHCHD4 | 4549 | 0.981 | 0.5123 | Yes | ||
| 62 | ATP11B | 4589 | 0.967 | 0.5151 | Yes | ||
| 63 | NFAT5 | 4779 | 0.904 | 0.5094 | No | ||
| 64 | ENTPD5 | 4916 | 0.861 | 0.5063 | No | ||
| 65 | POU4F2 | 5390 | 0.714 | 0.4844 | No | ||
| 66 | LRRC4 | 5462 | 0.691 | 0.4840 | No | ||
| 67 | TEAD1 | 5982 | 0.542 | 0.4586 | No | ||
| 68 | WTAP | 6211 | 0.476 | 0.4487 | No | ||
| 69 | AUH | 6696 | 0.346 | 0.4243 | No | ||
| 70 | TGFA | 7154 | 0.225 | 0.4007 | No | ||
| 71 | QSCN6L1 | 7222 | 0.209 | 0.3982 | No | ||
| 72 | EBF3 | 7389 | 0.168 | 0.3900 | No | ||
| 73 | BET1 | 8565 | -0.130 | 0.3272 | No | ||
| 74 | FBN2 | 8883 | -0.206 | 0.3111 | No | ||
| 75 | MYLIP | 8895 | -0.208 | 0.3115 | No | ||
| 76 | NRK | 9137 | -0.266 | 0.2998 | No | ||
| 77 | RCN1 | 9487 | -0.337 | 0.2827 | No | ||
| 78 | SEMA6D | 10187 | -0.506 | 0.2474 | No | ||
| 79 | LPP | 10333 | -0.541 | 0.2423 | No | ||
| 80 | STX12 | 10577 | -0.591 | 0.2321 | No | ||
| 81 | GPM6A | 10842 | -0.651 | 0.2211 | No | ||
| 82 | EIF3S1 | 10848 | -0.652 | 0.2241 | No | ||
| 83 | TMOD1 | 10874 | -0.658 | 0.2261 | No | ||
| 84 | DPF2 | 11174 | -0.719 | 0.2135 | No | ||
| 85 | POU2F3 | 11361 | -0.760 | 0.2073 | No | ||
| 86 | NFKB1 | 11453 | -0.779 | 0.2063 | No | ||
| 87 | CXCL12 | 11524 | -0.791 | 0.2064 | No | ||
| 88 | ZIC1 | 11927 | -0.874 | 0.1891 | No | ||
| 89 | MCFD2 | 12432 | -0.974 | 0.1667 | No | ||
| 90 | CRK | 12483 | -0.986 | 0.1690 | No | ||
| 91 | SLC4A4 | 12532 | -0.996 | 0.1714 | No | ||
| 92 | KCNJ3 | 12753 | -1.042 | 0.1647 | No | ||
| 93 | FBN1 | 12778 | -1.047 | 0.1686 | No | ||
| 94 | CDKN1B | 12982 | -1.090 | 0.1631 | No | ||
| 95 | RAB11FIP2 | 13008 | -1.094 | 0.1672 | No | ||
| 96 | STK35 | 13143 | -1.121 | 0.1656 | No | ||
| 97 | OSBP | 13654 | -1.244 | 0.1443 | No | ||
| 98 | HOXB5 | 13672 | -1.250 | 0.1496 | No | ||
| 99 | SEPT3 | 13839 | -1.292 | 0.1471 | No | ||
| 100 | HOXA1 | 14116 | -1.357 | 0.1390 | No | ||
| 101 | MAP3K3 | 14527 | -1.464 | 0.1242 | No | ||
| 102 | STAT5B | 14749 | -1.521 | 0.1198 | No | ||
| 103 | PAK7 | 14815 | -1.536 | 0.1240 | No | ||
| 104 | TMEM16D | 14977 | -1.587 | 0.1232 | No | ||
| 105 | ANKRD27 | 15009 | -1.594 | 0.1295 | No | ||
| 106 | ACAT2 | 16098 | -1.983 | 0.0807 | No | ||
| 107 | EGR3 | 16768 | -2.302 | 0.0561 | No | ||
| 108 | PTK2B | 18292 | -4.044 | -0.0060 | No | ||
| 109 | SPSB4 | 18457 | -4.687 | 0.0086 | No |
