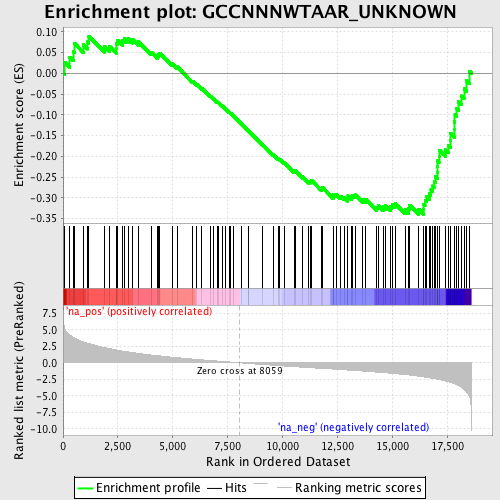
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCCNNNWTAAR_UNKNOWN |
| Enrichment Score (ES) | -0.33995995 |
| Normalized Enrichment Score (NES) | -1.487513 |
| Nominal p-value | 0.018621974 |
| FDR q-value | 0.41520044 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ITPR1 | 69 | 5.257 | 0.0271 | No | ||
| 2 | HPS3 | 303 | 4.205 | 0.0391 | No | ||
| 3 | PTMA | 470 | 3.836 | 0.0527 | No | ||
| 4 | RBMX | 509 | 3.763 | 0.0727 | No | ||
| 5 | SOCS2 | 916 | 3.168 | 0.0693 | No | ||
| 6 | ARID1A | 1116 | 2.949 | 0.0758 | No | ||
| 7 | ADAM12 | 1175 | 2.898 | 0.0897 | No | ||
| 8 | VCL | 1889 | 2.291 | 0.0646 | No | ||
| 9 | SOX4 | 2131 | 2.128 | 0.0641 | No | ||
| 10 | UBE2D3 | 2421 | 1.941 | 0.0598 | No | ||
| 11 | FYN | 2423 | 1.940 | 0.0712 | No | ||
| 12 | ZADH2 | 2467 | 1.914 | 0.0801 | No | ||
| 13 | NFATC4 | 2692 | 1.780 | 0.0784 | No | ||
| 14 | XPO7 | 2781 | 1.727 | 0.0838 | No | ||
| 15 | FES | 2962 | 1.634 | 0.0836 | No | ||
| 16 | ITGA3 | 3159 | 1.541 | 0.0821 | No | ||
| 17 | MPP2 | 3422 | 1.422 | 0.0762 | No | ||
| 18 | PPARGC1A | 4010 | 1.174 | 0.0514 | No | ||
| 19 | ERG | 4299 | 1.070 | 0.0421 | No | ||
| 20 | APBA2BP | 4351 | 1.049 | 0.0455 | No | ||
| 21 | SHH | 4400 | 1.033 | 0.0490 | No | ||
| 22 | ATOH1 | 4977 | 0.842 | 0.0228 | No | ||
| 23 | MITF | 5192 | 0.775 | 0.0158 | No | ||
| 24 | KCTD15 | 5910 | 0.566 | -0.0196 | No | ||
| 25 | EML4 | 6090 | 0.510 | -0.0263 | No | ||
| 26 | ISL1 | 6328 | 0.444 | -0.0365 | No | ||
| 27 | OTP | 6703 | 0.345 | -0.0547 | No | ||
| 28 | NPTX1 | 6864 | 0.300 | -0.0615 | No | ||
| 29 | SLC5A7 | 7028 | 0.255 | -0.0688 | No | ||
| 30 | PHEX | 7066 | 0.247 | -0.0694 | No | ||
| 31 | TGFB3 | 7257 | 0.201 | -0.0785 | No | ||
| 32 | EN2 | 7398 | 0.165 | -0.0851 | No | ||
| 33 | HOXA4 | 7565 | 0.125 | -0.0933 | No | ||
| 34 | JPH4 | 7647 | 0.104 | -0.0971 | No | ||
| 35 | NPR3 | 7759 | 0.077 | -0.1026 | No | ||
| 36 | SFXN5 | 8133 | -0.015 | -0.1227 | No | ||
| 37 | OTX1 | 8430 | -0.095 | -0.1381 | No | ||
| 38 | ADAMTS5 | 8444 | -0.100 | -0.1382 | No | ||
| 39 | MMP9 | 9089 | -0.256 | -0.1715 | No | ||
| 40 | KCNH3 | 9573 | -0.359 | -0.1955 | No | ||
| 41 | MEF2D | 9812 | -0.420 | -0.2059 | No | ||
| 42 | ABCA1 | 9869 | -0.432 | -0.2064 | No | ||
| 43 | SFTPC | 10089 | -0.485 | -0.2153 | No | ||
| 44 | DAB1 | 10530 | -0.582 | -0.2357 | No | ||
| 45 | PDGFB | 10575 | -0.591 | -0.2346 | No | ||
| 46 | TRIB2 | 10916 | -0.667 | -0.2491 | No | ||
| 47 | IMPDH2 | 11199 | -0.725 | -0.2601 | No | ||
| 48 | BHLHB3 | 11277 | -0.741 | -0.2599 | No | ||
| 49 | FGF9 | 11311 | -0.747 | -0.2573 | No | ||
| 50 | MID1 | 11775 | -0.841 | -0.2773 | No | ||
| 51 | HTR2C | 11819 | -0.852 | -0.2747 | No | ||
| 52 | COCH | 12307 | -0.949 | -0.2954 | No | ||
| 53 | RFX4 | 12334 | -0.956 | -0.2912 | No | ||
| 54 | ASGR1 | 12450 | -0.979 | -0.2917 | No | ||
| 55 | PHOX2B | 12652 | -1.023 | -0.2966 | No | ||
| 56 | GDNF | 12810 | -1.052 | -0.2989 | No | ||
| 57 | BCL9 | 12974 | -1.087 | -0.3013 | No | ||
| 58 | CDKN1B | 12982 | -1.090 | -0.2953 | No | ||
| 59 | STK35 | 13143 | -1.121 | -0.2974 | No | ||
| 60 | MSX1 | 13198 | -1.135 | -0.2936 | No | ||
| 61 | NDRG2 | 13310 | -1.160 | -0.2928 | No | ||
| 62 | LRP5 | 13660 | -1.246 | -0.3044 | No | ||
| 63 | RAB33A | 13798 | -1.282 | -0.3042 | No | ||
| 64 | HMGA2 | 14303 | -1.407 | -0.3232 | No | ||
| 65 | NFIL3 | 14358 | -1.420 | -0.3178 | No | ||
| 66 | H2AFX | 14586 | -1.480 | -0.3214 | No | ||
| 67 | FOXP2 | 14704 | -1.508 | -0.3189 | No | ||
| 68 | SOX5 | 14917 | -1.569 | -0.3211 | No | ||
| 69 | SLC25A10 | 14989 | -1.589 | -0.3157 | No | ||
| 70 | KLC2 | 15129 | -1.631 | -0.3136 | No | ||
| 71 | ABCC5 | 15596 | -1.773 | -0.3284 | Yes | ||
| 72 | DPF3 | 15754 | -1.836 | -0.3261 | Yes | ||
| 73 | TBCC | 15800 | -1.856 | -0.3176 | Yes | ||
| 74 | PEX5 | 16200 | -2.026 | -0.3273 | Yes | ||
| 75 | ATP5G2 | 16435 | -2.128 | -0.3275 | Yes | ||
| 76 | FHL3 | 16443 | -2.131 | -0.3154 | Yes | ||
| 77 | ELAVL4 | 16496 | -2.156 | -0.3055 | Yes | ||
| 78 | NKX2-5 | 16567 | -2.195 | -0.2965 | Yes | ||
| 79 | HOXB8 | 16694 | -2.261 | -0.2900 | Yes | ||
| 80 | PTPRG | 16765 | -2.301 | -0.2803 | Yes | ||
| 81 | HOXA10 | 16854 | -2.351 | -0.2713 | Yes | ||
| 82 | VAX1 | 16909 | -2.385 | -0.2602 | Yes | ||
| 83 | LDHD | 16970 | -2.430 | -0.2492 | Yes | ||
| 84 | SLC39A5 | 17055 | -2.481 | -0.2392 | Yes | ||
| 85 | CDKN2C | 17060 | -2.484 | -0.2249 | Yes | ||
| 86 | EGR1 | 17072 | -2.490 | -0.2109 | Yes | ||
| 87 | CAMK2D | 17135 | -2.535 | -0.1994 | Yes | ||
| 88 | RPL10 | 17145 | -2.541 | -0.1850 | Yes | ||
| 89 | MMP24 | 17435 | -2.762 | -0.1844 | Yes | ||
| 90 | CTDSP1 | 17552 | -2.863 | -0.1739 | Yes | ||
| 91 | GNAQ | 17642 | -2.964 | -0.1613 | Yes | ||
| 92 | INPPL1 | 17649 | -2.969 | -0.1442 | Yes | ||
| 93 | ETV5 | 17825 | -3.153 | -0.1352 | Yes | ||
| 94 | ETV6 | 17829 | -3.156 | -0.1169 | Yes | ||
| 95 | EGFL6 | 17860 | -3.206 | -0.0997 | Yes | ||
| 96 | VASP | 17934 | -3.311 | -0.0842 | Yes | ||
| 97 | CCBL1 | 18017 | -3.444 | -0.0685 | Yes | ||
| 98 | BRUNOL4 | 18160 | -3.697 | -0.0545 | Yes | ||
| 99 | GGA1 | 18274 | -3.990 | -0.0372 | Yes | ||
| 100 | GRASP | 18395 | -4.399 | -0.0179 | Yes | ||
| 101 | RPP21 | 18520 | -5.078 | 0.0052 | Yes |
