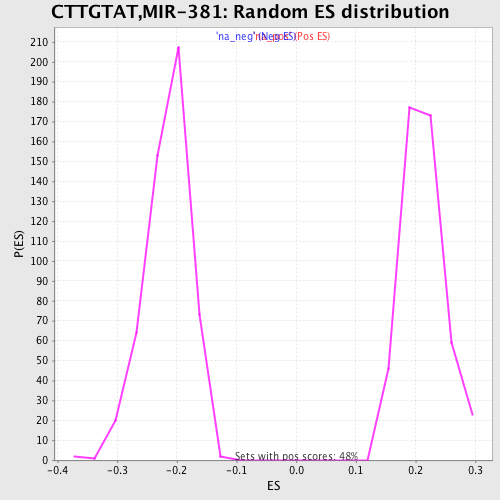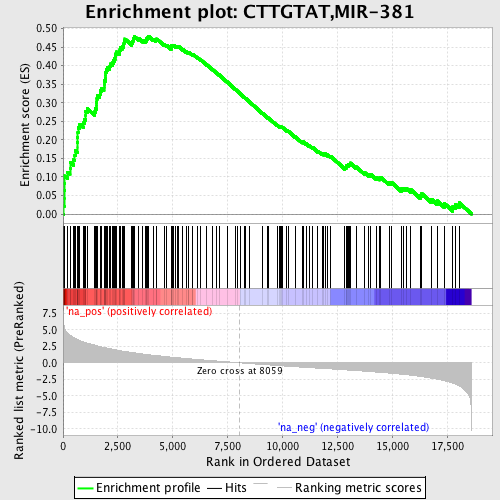
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CTTGTAT,MIR-381 |
| Enrichment Score (ES) | 0.47973973 |
| Normalized Enrichment Score (NES) | 2.26055 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APPBP2 | 21 | 5.998 | 0.0223 | Yes | ||
| 2 | GTF2I | 47 | 5.525 | 0.0425 | Yes | ||
| 3 | ZFYVE20 | 51 | 5.476 | 0.0638 | Yes | ||
| 4 | SATB1 | 70 | 5.244 | 0.0833 | Yes | ||
| 5 | BRD7 | 80 | 5.133 | 0.1028 | Yes | ||
| 6 | EIF4G3 | 211 | 4.479 | 0.1133 | Yes | ||
| 7 | CHD4 | 314 | 4.175 | 0.1241 | Yes | ||
| 8 | BCL11B | 324 | 4.156 | 0.1398 | Yes | ||
| 9 | CXCR4 | 479 | 3.818 | 0.1464 | Yes | ||
| 10 | ROD1 | 533 | 3.714 | 0.1580 | Yes | ||
| 11 | CNOT7 | 557 | 3.681 | 0.1712 | Yes | ||
| 12 | SLC38A2 | 645 | 3.552 | 0.1804 | Yes | ||
| 13 | KHDRBS1 | 646 | 3.552 | 0.1942 | Yes | ||
| 14 | EIF4G2 | 660 | 3.525 | 0.2073 | Yes | ||
| 15 | SLMAP | 674 | 3.502 | 0.2203 | Yes | ||
| 16 | HIPK1 | 686 | 3.486 | 0.2333 | Yes | ||
| 17 | INSIG1 | 759 | 3.367 | 0.2425 | Yes | ||
| 18 | HBP1 | 926 | 3.155 | 0.2459 | Yes | ||
| 19 | ZMYND11 | 965 | 3.111 | 0.2560 | Yes | ||
| 20 | RAP2C | 1021 | 3.061 | 0.2650 | Yes | ||
| 21 | APRIN | 1027 | 3.058 | 0.2766 | Yes | ||
| 22 | UBR1 | 1100 | 2.978 | 0.2844 | Yes | ||
| 23 | ETF1 | 1448 | 2.649 | 0.2759 | Yes | ||
| 24 | ASXL2 | 1479 | 2.617 | 0.2845 | Yes | ||
| 25 | CEBPA | 1522 | 2.578 | 0.2923 | Yes | ||
| 26 | GJA1 | 1527 | 2.575 | 0.3022 | Yes | ||
| 27 | PDAP1 | 1541 | 2.562 | 0.3115 | Yes | ||
| 28 | EIF3S10 | 1569 | 2.537 | 0.3199 | Yes | ||
| 29 | CSTF3 | 1681 | 2.449 | 0.3235 | Yes | ||
| 30 | RNF111 | 1694 | 2.437 | 0.3324 | Yes | ||
| 31 | NIN | 1734 | 2.410 | 0.3397 | Yes | ||
| 32 | SYNCRIP | 1864 | 2.313 | 0.3417 | Yes | ||
| 33 | EIF1 | 1867 | 2.308 | 0.3506 | Yes | ||
| 34 | RBM16 | 1874 | 2.302 | 0.3593 | Yes | ||
| 35 | KPNA3 | 1922 | 2.272 | 0.3656 | Yes | ||
| 36 | NLK | 1937 | 2.261 | 0.3737 | Yes | ||
| 37 | WEE1 | 1943 | 2.258 | 0.3822 | Yes | ||
| 38 | ROCK2 | 1973 | 2.242 | 0.3894 | Yes | ||
| 39 | SEMA3C | 2011 | 2.215 | 0.3961 | Yes | ||
| 40 | SOX4 | 2131 | 2.128 | 0.3980 | Yes | ||
| 41 | STAG1 | 2159 | 2.109 | 0.4047 | Yes | ||
| 42 | NDUFAB1 | 2254 | 2.050 | 0.4077 | Yes | ||
| 43 | KLF4 | 2293 | 2.019 | 0.4135 | Yes | ||
| 44 | YTHDF1 | 2334 | 1.989 | 0.4191 | Yes | ||
| 45 | THRAP2 | 2369 | 1.968 | 0.4249 | Yes | ||
| 46 | ANKRD50 | 2388 | 1.957 | 0.4316 | Yes | ||
| 47 | KCTD3 | 2427 | 1.936 | 0.4371 | Yes | ||
| 48 | SLC6A8 | 2558 | 1.858 | 0.4373 | Yes | ||
| 49 | DDX6 | 2589 | 1.848 | 0.4429 | Yes | ||
| 50 | ITCH | 2613 | 1.837 | 0.4489 | Yes | ||
| 51 | PSIP1 | 2689 | 1.781 | 0.4518 | Yes | ||
| 52 | AZIN1 | 2753 | 1.740 | 0.4551 | Yes | ||
| 53 | PHF2 | 2771 | 1.731 | 0.4610 | Yes | ||
| 54 | XPO7 | 2781 | 1.727 | 0.4672 | Yes | ||
| 55 | PAPOLG | 2818 | 1.709 | 0.4720 | Yes | ||
| 56 | HNRPH2 | 3134 | 1.551 | 0.4610 | Yes | ||
| 57 | GAD1 | 3169 | 1.537 | 0.4651 | Yes | ||
| 58 | R3HDM1 | 3190 | 1.527 | 0.4700 | Yes | ||
| 59 | SNRK | 3215 | 1.517 | 0.4746 | Yes | ||
| 60 | RNF144 | 3231 | 1.512 | 0.4797 | Yes | ||
| 61 | RPS6KA3 | 3437 | 1.415 | 0.4742 | No | ||
| 62 | KIF1B | 3631 | 1.327 | 0.4689 | No | ||
| 63 | THRAP1 | 3741 | 1.280 | 0.4680 | No | ||
| 64 | ATRN | 3786 | 1.265 | 0.4706 | No | ||
| 65 | ARID4B | 3811 | 1.250 | 0.4741 | No | ||
| 66 | ACSL3 | 3861 | 1.230 | 0.4763 | No | ||
| 67 | CGGBP1 | 3892 | 1.218 | 0.4794 | No | ||
| 68 | MTSS1 | 4119 | 1.132 | 0.4716 | No | ||
| 69 | ZFAND3 | 4233 | 1.094 | 0.4698 | No | ||
| 70 | QKI | 4270 | 1.078 | 0.4720 | No | ||
| 71 | ELOVL7 | 4613 | 0.959 | 0.4573 | No | ||
| 72 | MBNL2 | 4723 | 0.923 | 0.4550 | No | ||
| 73 | OSBPL3 | 4924 | 0.860 | 0.4475 | No | ||
| 74 | PURG | 4932 | 0.858 | 0.4505 | No | ||
| 75 | B3GNT5 | 4936 | 0.856 | 0.4536 | No | ||
| 76 | GRM1 | 4967 | 0.844 | 0.4553 | No | ||
| 77 | SLC24A3 | 5041 | 0.822 | 0.4546 | No | ||
| 78 | BHLHB5 | 5144 | 0.789 | 0.4521 | No | ||
| 79 | MITF | 5192 | 0.775 | 0.4526 | No | ||
| 80 | NFKBIA | 5250 | 0.761 | 0.4525 | No | ||
| 81 | LRRC4 | 5462 | 0.691 | 0.4438 | No | ||
| 82 | JAG2 | 5625 | 0.638 | 0.4375 | No | ||
| 83 | PAPOLA | 5716 | 0.615 | 0.4350 | No | ||
| 84 | PBEF1 | 5734 | 0.611 | 0.4365 | No | ||
| 85 | SSH2 | 5898 | 0.569 | 0.4299 | No | ||
| 86 | KCTD15 | 5910 | 0.566 | 0.4315 | No | ||
| 87 | FOXF2 | 6118 | 0.503 | 0.4223 | No | ||
| 88 | LZTS2 | 6254 | 0.466 | 0.4168 | No | ||
| 89 | CCNT2 | 6551 | 0.387 | 0.4023 | No | ||
| 90 | UBE4A | 6811 | 0.319 | 0.3895 | No | ||
| 91 | GAS2 | 6969 | 0.273 | 0.3821 | No | ||
| 92 | JPH1 | 7106 | 0.236 | 0.3756 | No | ||
| 93 | BTBD7 | 7471 | 0.147 | 0.3565 | No | ||
| 94 | BIRC6 | 7864 | 0.051 | 0.3355 | No | ||
| 95 | RPL35A | 7956 | 0.025 | 0.3306 | No | ||
| 96 | UBE2E2 | 8091 | -0.006 | 0.3234 | No | ||
| 97 | RHOQ | 8263 | -0.051 | 0.3143 | No | ||
| 98 | ZFPM2 | 8298 | -0.059 | 0.3127 | No | ||
| 99 | PARD6B | 8504 | -0.116 | 0.3021 | No | ||
| 100 | RNF2 | 9081 | -0.255 | 0.2719 | No | ||
| 101 | EPHA3 | 9332 | -0.305 | 0.2595 | No | ||
| 102 | TBC1D15 | 9377 | -0.314 | 0.2584 | No | ||
| 103 | ST8SIA2 | 9765 | -0.406 | 0.2390 | No | ||
| 104 | ABCA1 | 9869 | -0.432 | 0.2351 | No | ||
| 105 | PMP22 | 9902 | -0.442 | 0.2351 | No | ||
| 106 | SRRM1 | 9937 | -0.449 | 0.2350 | No | ||
| 107 | PNRC1 | 9991 | -0.462 | 0.2340 | No | ||
| 108 | SEMA6D | 10187 | -0.506 | 0.2254 | No | ||
| 109 | CLCF1 | 10253 | -0.519 | 0.2239 | No | ||
| 110 | STX12 | 10577 | -0.591 | 0.2087 | No | ||
| 111 | SIPA1L2 | 10896 | -0.663 | 0.1941 | No | ||
| 112 | PDGFC | 10933 | -0.669 | 0.1947 | No | ||
| 113 | CRSP2 | 11079 | -0.698 | 0.1896 | No | ||
| 114 | SHANK2 | 11236 | -0.733 | 0.1840 | No | ||
| 115 | NAP1L5 | 11386 | -0.766 | 0.1790 | No | ||
| 116 | HNRPR | 11614 | -0.811 | 0.1698 | No | ||
| 117 | ACVR2A | 11808 | -0.850 | 0.1627 | No | ||
| 118 | PHF15 | 11875 | -0.863 | 0.1625 | No | ||
| 119 | ZCCHC11 | 11941 | -0.877 | 0.1624 | No | ||
| 120 | EPHA4 | 12057 | -0.898 | 0.1597 | No | ||
| 121 | NRXN1 | 12208 | -0.928 | 0.1552 | No | ||
| 122 | CACNA1C | 12845 | -1.060 | 0.1249 | No | ||
| 123 | VSNL1 | 12898 | -1.071 | 0.1263 | No | ||
| 124 | NR5A2 | 12904 | -1.072 | 0.1302 | No | ||
| 125 | RNF13 | 12954 | -1.085 | 0.1318 | No | ||
| 126 | RAB11FIP2 | 13008 | -1.094 | 0.1332 | No | ||
| 127 | MDS1 | 13063 | -1.105 | 0.1346 | No | ||
| 128 | BAT2 | 13085 | -1.108 | 0.1378 | No | ||
| 129 | COL3A1 | 13367 | -1.173 | 0.1271 | No | ||
| 130 | TRIM63 | 13737 | -1.264 | 0.1121 | No | ||
| 131 | BCL11A | 13924 | -1.312 | 0.1071 | No | ||
| 132 | NFIA | 14031 | -1.334 | 0.1066 | No | ||
| 133 | WNT5A | 14294 | -1.403 | 0.0979 | No | ||
| 134 | FLRT3 | 14407 | -1.434 | 0.0974 | No | ||
| 135 | ETS1 | 14475 | -1.449 | 0.0995 | No | ||
| 136 | GORASP2 | 14856 | -1.549 | 0.0849 | No | ||
| 137 | TMEM16D | 14977 | -1.587 | 0.0846 | No | ||
| 138 | UBE3A | 15409 | -1.719 | 0.0680 | No | ||
| 139 | GRM8 | 15496 | -1.744 | 0.0702 | No | ||
| 140 | KLF12 | 15631 | -1.785 | 0.0699 | No | ||
| 141 | RALGPS1 | 15842 | -1.875 | 0.0659 | No | ||
| 142 | UBN1 | 16281 | -2.057 | 0.0502 | No | ||
| 143 | ACTG1 | 16348 | -2.090 | 0.0548 | No | ||
| 144 | LPHN1 | 16795 | -2.322 | 0.0397 | No | ||
| 145 | RPS6KB1 | 17050 | -2.477 | 0.0356 | No | ||
| 146 | AGPAT3 | 17368 | -2.716 | 0.0291 | No | ||
| 147 | DHX40 | 17765 | -3.092 | 0.0197 | No | ||
| 148 | GFRA1 | 17875 | -3.221 | 0.0264 | No | ||
| 149 | DLC1 | 18054 | -3.505 | 0.0304 | No |