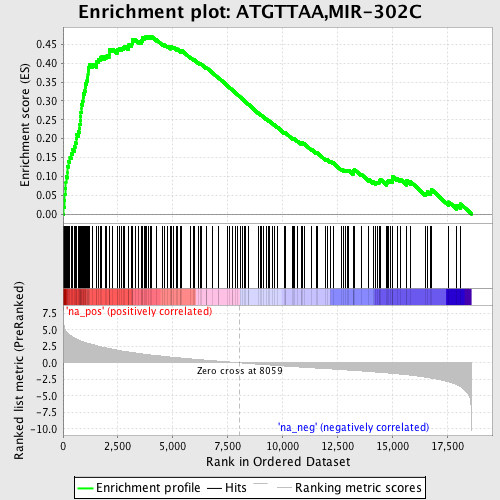
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ATGTTAA,MIR-302C |
| Enrichment Score (ES) | 0.47175035 |
| Normalized Enrichment Score (NES) | 2.2878768 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UBQLN1 | 19 | 6.067 | 0.0197 | Yes | ||
| 2 | H3F3B | 65 | 5.308 | 0.0355 | Yes | ||
| 3 | PPP3R1 | 77 | 5.168 | 0.0525 | Yes | ||
| 4 | METAP1 | 91 | 5.017 | 0.0690 | Yes | ||
| 5 | NFYB | 128 | 4.777 | 0.0834 | Yes | ||
| 6 | RAB14 | 132 | 4.754 | 0.0995 | Yes | ||
| 7 | ACBD3 | 204 | 4.515 | 0.1111 | Yes | ||
| 8 | RNF11 | 208 | 4.505 | 0.1264 | Yes | ||
| 9 | SMAD4 | 241 | 4.376 | 0.1396 | Yes | ||
| 10 | ZFP36L1 | 311 | 4.187 | 0.1502 | Yes | ||
| 11 | ACACA | 383 | 4.013 | 0.1601 | Yes | ||
| 12 | SLC35A1 | 421 | 3.920 | 0.1715 | Yes | ||
| 13 | SPOP | 529 | 3.732 | 0.1784 | Yes | ||
| 14 | RAP1B | 546 | 3.696 | 0.1902 | Yes | ||
| 15 | SENP6 | 595 | 3.626 | 0.2000 | Yes | ||
| 16 | RNF6 | 607 | 3.612 | 0.2118 | Yes | ||
| 17 | FOXP1 | 705 | 3.457 | 0.2184 | Yes | ||
| 18 | PTP4A2 | 755 | 3.372 | 0.2272 | Yes | ||
| 19 | PARP1 | 762 | 3.364 | 0.2384 | Yes | ||
| 20 | MAPRE1 | 785 | 3.327 | 0.2486 | Yes | ||
| 21 | MBNL1 | 795 | 3.320 | 0.2595 | Yes | ||
| 22 | PPP2R5E | 809 | 3.301 | 0.2701 | Yes | ||
| 23 | SFRS2 | 836 | 3.269 | 0.2799 | Yes | ||
| 24 | DDX3X | 840 | 3.265 | 0.2909 | Yes | ||
| 25 | SFRS5 | 879 | 3.212 | 0.2998 | Yes | ||
| 26 | HNRPH1 | 909 | 3.176 | 0.3091 | Yes | ||
| 27 | PPP1R12A | 918 | 3.166 | 0.3195 | Yes | ||
| 28 | TOB1 | 982 | 3.095 | 0.3267 | Yes | ||
| 29 | HNRPA2B1 | 1015 | 3.064 | 0.3354 | Yes | ||
| 30 | NFIX | 1025 | 3.058 | 0.3454 | Yes | ||
| 31 | EPC2 | 1078 | 3.007 | 0.3529 | Yes | ||
| 32 | EIF4A2 | 1098 | 2.986 | 0.3620 | Yes | ||
| 33 | IMMT | 1133 | 2.934 | 0.3702 | Yes | ||
| 34 | ABR | 1136 | 2.931 | 0.3802 | Yes | ||
| 35 | RNF138 | 1159 | 2.911 | 0.3889 | Yes | ||
| 36 | TMEM33 | 1190 | 2.890 | 0.3972 | Yes | ||
| 37 | SIRT1 | 1352 | 2.740 | 0.3978 | Yes | ||
| 38 | AMMECR1 | 1519 | 2.581 | 0.3977 | Yes | ||
| 39 | TBL1X | 1534 | 2.569 | 0.4057 | Yes | ||
| 40 | USP1 | 1620 | 2.495 | 0.4096 | Yes | ||
| 41 | GABPA | 1689 | 2.441 | 0.4143 | Yes | ||
| 42 | MAGI1 | 1768 | 2.386 | 0.4182 | Yes | ||
| 43 | ATP6V1A | 1909 | 2.279 | 0.4184 | Yes | ||
| 44 | KIF4A | 1999 | 2.223 | 0.4212 | Yes | ||
| 45 | IL13RA1 | 2094 | 2.164 | 0.4235 | Yes | ||
| 46 | ZIC3 | 2100 | 2.160 | 0.4306 | Yes | ||
| 47 | CYFIP2 | 2119 | 2.138 | 0.4370 | Yes | ||
| 48 | FBXO33 | 2248 | 2.054 | 0.4371 | Yes | ||
| 49 | ATP6V1B2 | 2461 | 1.916 | 0.4321 | Yes | ||
| 50 | ZNF319 | 2487 | 1.902 | 0.4373 | Yes | ||
| 51 | SAMD8 | 2552 | 1.862 | 0.4402 | Yes | ||
| 52 | PANK3 | 2677 | 1.788 | 0.4396 | Yes | ||
| 53 | AZIN1 | 2753 | 1.740 | 0.4415 | Yes | ||
| 54 | DMRT2 | 2810 | 1.712 | 0.4443 | Yes | ||
| 55 | EDNRB | 2980 | 1.625 | 0.4407 | Yes | ||
| 56 | DNAJB6 | 2988 | 1.620 | 0.4459 | Yes | ||
| 57 | MATR3 | 2999 | 1.615 | 0.4508 | Yes | ||
| 58 | SON | 3127 | 1.555 | 0.4493 | Yes | ||
| 59 | NRIP1 | 3141 | 1.549 | 0.4539 | Yes | ||
| 60 | RAI1 | 3160 | 1.540 | 0.4582 | Yes | ||
| 61 | GAD1 | 3169 | 1.537 | 0.4630 | Yes | ||
| 62 | ATP10A | 3286 | 1.486 | 0.4618 | Yes | ||
| 63 | SSX2IP | 3445 | 1.411 | 0.4580 | Yes | ||
| 64 | FKBP4 | 3559 | 1.363 | 0.4566 | Yes | ||
| 65 | PDE4D | 3575 | 1.357 | 0.4604 | Yes | ||
| 66 | PCDH9 | 3616 | 1.333 | 0.4628 | Yes | ||
| 67 | ASF1A | 3617 | 1.333 | 0.4674 | Yes | ||
| 68 | SOX11 | 3704 | 1.297 | 0.4671 | Yes | ||
| 69 | THRAP1 | 3741 | 1.280 | 0.4696 | Yes | ||
| 70 | USP33 | 3805 | 1.254 | 0.4704 | Yes | ||
| 71 | CGGBP1 | 3892 | 1.218 | 0.4700 | Yes | ||
| 72 | NUMB | 3975 | 1.190 | 0.4696 | Yes | ||
| 73 | PPARGC1A | 4010 | 1.174 | 0.4718 | Yes | ||
| 74 | NR3C2 | 4254 | 1.088 | 0.4623 | No | ||
| 75 | SNAP91 | 4542 | 0.984 | 0.4501 | No | ||
| 76 | BHLHB2 | 4633 | 0.952 | 0.4485 | No | ||
| 77 | NFAT5 | 4779 | 0.904 | 0.4437 | No | ||
| 78 | DDX3Y | 4895 | 0.868 | 0.4404 | No | ||
| 79 | STRN | 4930 | 0.858 | 0.4415 | No | ||
| 80 | B3GNT5 | 4936 | 0.856 | 0.4442 | No | ||
| 81 | FBXL3 | 5013 | 0.828 | 0.4429 | No | ||
| 82 | PLEKHA5 | 5179 | 0.779 | 0.4366 | No | ||
| 83 | TNFSF11 | 5208 | 0.773 | 0.4378 | No | ||
| 84 | UNC5D | 5368 | 0.721 | 0.4316 | No | ||
| 85 | GOLPH3 | 5371 | 0.719 | 0.4340 | No | ||
| 86 | PCF11 | 5416 | 0.708 | 0.4340 | No | ||
| 87 | DLX1 | 5811 | 0.592 | 0.4146 | No | ||
| 88 | FMN2 | 5963 | 0.548 | 0.4083 | No | ||
| 89 | TEAD1 | 5982 | 0.542 | 0.4092 | No | ||
| 90 | PPP5C | 6172 | 0.486 | 0.4006 | No | ||
| 91 | TTC7B | 6239 | 0.470 | 0.3987 | No | ||
| 92 | FBXO21 | 6299 | 0.450 | 0.3970 | No | ||
| 93 | ACVR1 | 6525 | 0.393 | 0.3861 | No | ||
| 94 | ZCCHC16 | 6532 | 0.391 | 0.3872 | No | ||
| 95 | CCNT2 | 6551 | 0.387 | 0.3875 | No | ||
| 96 | GPRASP2 | 6805 | 0.321 | 0.3749 | No | ||
| 97 | PCBP1 | 7086 | 0.242 | 0.3605 | No | ||
| 98 | USP15 | 7101 | 0.236 | 0.3606 | No | ||
| 99 | UBB | 7493 | 0.142 | 0.3399 | No | ||
| 100 | HBEGF | 7600 | 0.116 | 0.3345 | No | ||
| 101 | NEUROG2 | 7703 | 0.089 | 0.3293 | No | ||
| 102 | MTMR6 | 7734 | 0.084 | 0.3279 | No | ||
| 103 | SBF2 | 7835 | 0.060 | 0.3227 | No | ||
| 104 | PPARA | 7960 | 0.023 | 0.3161 | No | ||
| 105 | SP4 | 8066 | -0.002 | 0.3104 | No | ||
| 106 | LRCH2 | 8197 | -0.032 | 0.3035 | No | ||
| 107 | KCTD8 | 8283 | -0.055 | 0.2990 | No | ||
| 108 | ZFPM2 | 8298 | -0.059 | 0.2985 | No | ||
| 109 | YY1 | 8443 | -0.100 | 0.2910 | No | ||
| 110 | NUP153 | 8465 | -0.107 | 0.2902 | No | ||
| 111 | DAZAP1 | 8886 | -0.207 | 0.2682 | No | ||
| 112 | MYT1 | 8989 | -0.228 | 0.2634 | No | ||
| 113 | CBLN2 | 9031 | -0.240 | 0.2620 | No | ||
| 114 | HOXA13 | 9119 | -0.262 | 0.2582 | No | ||
| 115 | GABRB3 | 9250 | -0.288 | 0.2521 | No | ||
| 116 | OTX2 | 9341 | -0.307 | 0.2483 | No | ||
| 117 | GABRA2 | 9388 | -0.317 | 0.2469 | No | ||
| 118 | MYO1B | 9537 | -0.352 | 0.2401 | No | ||
| 119 | SOX3 | 9623 | -0.372 | 0.2368 | No | ||
| 120 | UBE2V2 | 9775 | -0.408 | 0.2300 | No | ||
| 121 | TBR1 | 10086 | -0.483 | 0.2148 | No | ||
| 122 | DACH1 | 10125 | -0.494 | 0.2144 | No | ||
| 123 | KHDRBS3 | 10135 | -0.496 | 0.2156 | No | ||
| 124 | GPC4 | 10461 | -0.567 | 0.2000 | No | ||
| 125 | PTPN3 | 10517 | -0.579 | 0.1990 | No | ||
| 126 | C1QTNF7 | 10544 | -0.585 | 0.1996 | No | ||
| 127 | CPEB2 | 10697 | -0.619 | 0.1934 | No | ||
| 128 | EIF3S1 | 10848 | -0.652 | 0.1875 | No | ||
| 129 | HDHD2 | 10857 | -0.655 | 0.1893 | No | ||
| 130 | SIPA1L2 | 10896 | -0.663 | 0.1895 | No | ||
| 131 | FOXA1 | 11001 | -0.681 | 0.1862 | No | ||
| 132 | NPY2R | 11325 | -0.749 | 0.1713 | No | ||
| 133 | CSNK1D | 11534 | -0.794 | 0.1627 | No | ||
| 134 | CREM | 11594 | -0.807 | 0.1623 | No | ||
| 135 | ST6GALNAC3 | 11956 | -0.879 | 0.1457 | No | ||
| 136 | FZD4 | 12041 | -0.896 | 0.1442 | No | ||
| 137 | TPD52 | 12181 | -0.922 | 0.1399 | No | ||
| 138 | ATP2C1 | 12306 | -0.949 | 0.1364 | No | ||
| 139 | B4GALT5 | 12707 | -1.034 | 0.1182 | No | ||
| 140 | KHDRBS2 | 12799 | -1.050 | 0.1169 | No | ||
| 141 | TLL1 | 12890 | -1.068 | 0.1157 | No | ||
| 142 | RNF13 | 12954 | -1.085 | 0.1160 | No | ||
| 143 | ZBTB4 | 13024 | -1.096 | 0.1160 | No | ||
| 144 | WNT3 | 13218 | -1.139 | 0.1094 | No | ||
| 145 | ARL4A | 13222 | -1.141 | 0.1131 | No | ||
| 146 | FMR1 | 13235 | -1.144 | 0.1164 | No | ||
| 147 | BTBD3 | 13287 | -1.156 | 0.1176 | No | ||
| 148 | RHEB | 13593 | -1.228 | 0.1053 | No | ||
| 149 | BCL11A | 13924 | -1.312 | 0.0919 | No | ||
| 150 | CSMD3 | 14128 | -1.359 | 0.0855 | No | ||
| 151 | FGFR2 | 14249 | -1.390 | 0.0837 | No | ||
| 152 | PCSK6 | 14320 | -1.410 | 0.0848 | No | ||
| 153 | LIFR | 14398 | -1.432 | 0.0855 | No | ||
| 154 | NRP2 | 14423 | -1.437 | 0.0891 | No | ||
| 155 | SH3D19 | 14474 | -1.449 | 0.0914 | No | ||
| 156 | MAK | 14744 | -1.520 | 0.0820 | No | ||
| 157 | BMP2 | 14766 | -1.524 | 0.0861 | No | ||
| 158 | PAK7 | 14815 | -1.536 | 0.0887 | No | ||
| 159 | ZMYND19 | 14911 | -1.567 | 0.0889 | No | ||
| 160 | PHACTR2 | 15014 | -1.596 | 0.0888 | No | ||
| 161 | PTPRE | 15023 | -1.599 | 0.0939 | No | ||
| 162 | SUV39H2 | 15026 | -1.600 | 0.0993 | No | ||
| 163 | SF3B1 | 15217 | -1.661 | 0.0946 | No | ||
| 164 | POU3F2 | 15369 | -1.706 | 0.0923 | No | ||
| 165 | LRRTM4 | 15654 | -1.794 | 0.0830 | No | ||
| 166 | GPHN | 15665 | -1.798 | 0.0886 | No | ||
| 167 | SLC30A8 | 15840 | -1.875 | 0.0856 | No | ||
| 168 | ADD3 | 16532 | -2.177 | 0.0556 | No | ||
| 169 | SPRY1 | 16601 | -2.215 | 0.0595 | No | ||
| 170 | CREB5 | 16764 | -2.299 | 0.0586 | No | ||
| 171 | HOXB3 | 16779 | -2.314 | 0.0657 | No | ||
| 172 | EHD3 | 17582 | -2.891 | 0.0321 | No | ||
| 173 | DAAM1 | 17943 | -3.325 | 0.0240 | No | ||
| 174 | WDTC1 | 18119 | -3.634 | 0.0269 | No |
