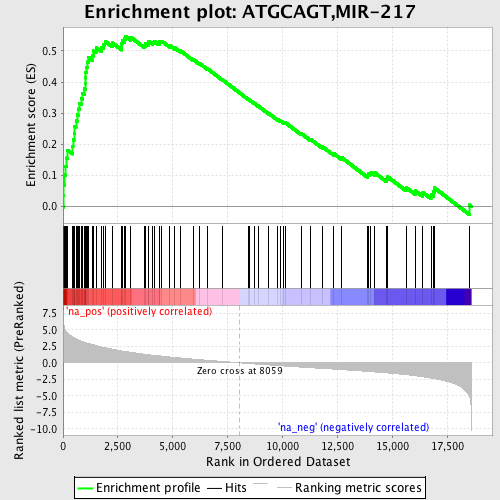
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | 0.5476785 |
| Normalized Enrichment Score (NES) | 2.308727 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APPBP2 | 21 | 5.998 | 0.0356 | Yes | ||
| 2 | PPM1D | 36 | 5.779 | 0.0701 | Yes | ||
| 3 | ZFYVE20 | 51 | 5.476 | 0.1029 | Yes | ||
| 4 | DYNC1LI2 | 104 | 4.942 | 0.1303 | Yes | ||
| 5 | IVNS1ABP | 152 | 4.681 | 0.1564 | Yes | ||
| 6 | NDEL1 | 194 | 4.555 | 0.1821 | Yes | ||
| 7 | MYEF2 | 433 | 3.898 | 0.1931 | Yes | ||
| 8 | EHMT1 | 452 | 3.857 | 0.2157 | Yes | ||
| 9 | UBL3 | 499 | 3.780 | 0.2363 | Yes | ||
| 10 | SENP5 | 534 | 3.714 | 0.2572 | Yes | ||
| 11 | KRAS | 590 | 3.637 | 0.2765 | Yes | ||
| 12 | SLC38A2 | 645 | 3.552 | 0.2953 | Yes | ||
| 13 | YAF2 | 697 | 3.470 | 0.3138 | Yes | ||
| 14 | PCNA | 752 | 3.379 | 0.3315 | Yes | ||
| 15 | SLC31A1 | 823 | 3.280 | 0.3478 | Yes | ||
| 16 | MAPK1 | 896 | 3.193 | 0.3635 | Yes | ||
| 17 | ATP1B1 | 962 | 3.116 | 0.3790 | Yes | ||
| 18 | HNRPA2B1 | 1015 | 3.064 | 0.3949 | Yes | ||
| 19 | RAP2C | 1021 | 3.061 | 0.4134 | Yes | ||
| 20 | APRIN | 1027 | 3.058 | 0.4318 | Yes | ||
| 21 | RTF1 | 1070 | 3.013 | 0.4480 | Yes | ||
| 22 | EIF4A2 | 1098 | 2.986 | 0.4648 | Yes | ||
| 23 | TLK2 | 1162 | 2.905 | 0.4792 | Yes | ||
| 24 | SIRT1 | 1352 | 2.740 | 0.4857 | Yes | ||
| 25 | MTPN | 1392 | 2.703 | 0.5002 | Yes | ||
| 26 | MSN | 1501 | 2.593 | 0.5102 | Yes | ||
| 27 | HNRPUL1 | 1736 | 2.408 | 0.5123 | Yes | ||
| 28 | STX6 | 1835 | 2.336 | 0.5213 | Yes | ||
| 29 | ATP6V1A | 1909 | 2.279 | 0.5313 | Yes | ||
| 30 | OGT | 2242 | 2.058 | 0.5260 | Yes | ||
| 31 | FN1 | 2668 | 1.796 | 0.5140 | Yes | ||
| 32 | KCTD9 | 2674 | 1.792 | 0.5247 | Yes | ||
| 33 | RIN2 | 2700 | 1.775 | 0.5342 | Yes | ||
| 34 | DMRT2 | 2810 | 1.712 | 0.5388 | Yes | ||
| 35 | EZH2 | 2839 | 1.693 | 0.5477 | Yes | ||
| 36 | NR4A2 | 3084 | 1.575 | 0.5442 | No | ||
| 37 | SOX11 | 3704 | 1.297 | 0.5187 | No | ||
| 38 | YWHAG | 3760 | 1.274 | 0.5235 | No | ||
| 39 | STRBP | 3886 | 1.220 | 0.5242 | No | ||
| 40 | TMSB4X | 3891 | 1.219 | 0.5315 | No | ||
| 41 | ATP11C | 4089 | 1.145 | 0.5279 | No | ||
| 42 | GDI2 | 4179 | 1.115 | 0.5299 | No | ||
| 43 | JOSD3 | 4378 | 1.038 | 0.5255 | No | ||
| 44 | BAI3 | 4396 | 1.034 | 0.5310 | No | ||
| 45 | DYRK1A | 4492 | 1.003 | 0.5320 | No | ||
| 46 | STAG2 | 4858 | 0.881 | 0.5177 | No | ||
| 47 | SLC39A10 | 5089 | 0.807 | 0.5102 | No | ||
| 48 | ZCCHC2 | 5348 | 0.728 | 0.5007 | No | ||
| 49 | ELOVL4 | 5948 | 0.551 | 0.4718 | No | ||
| 50 | STX1A | 6213 | 0.476 | 0.4605 | No | ||
| 51 | MAMDC2 | 6586 | 0.377 | 0.4427 | No | ||
| 52 | CUL5 | 7253 | 0.202 | 0.4080 | No | ||
| 53 | ADAMTS5 | 8444 | -0.100 | 0.3444 | No | ||
| 54 | POLG | 8505 | -0.116 | 0.3419 | No | ||
| 55 | FBXW11 | 8723 | -0.167 | 0.3312 | No | ||
| 56 | FBN2 | 8883 | -0.206 | 0.3239 | No | ||
| 57 | TBC1D15 | 9377 | -0.314 | 0.2992 | No | ||
| 58 | AKAP2 | 9791 | -0.412 | 0.2795 | No | ||
| 59 | SLITRK3 | 9892 | -0.438 | 0.2768 | No | ||
| 60 | DLGAP2 | 10066 | -0.478 | 0.2703 | No | ||
| 61 | DACH1 | 10125 | -0.494 | 0.2702 | No | ||
| 62 | GPM6A | 10842 | -0.651 | 0.2356 | No | ||
| 63 | A2BP1 | 11282 | -0.742 | 0.2165 | No | ||
| 64 | ACVR2A | 11808 | -0.850 | 0.1933 | No | ||
| 65 | PAQR3 | 12319 | -0.952 | 0.1716 | No | ||
| 66 | SEPT4 | 12685 | -1.030 | 0.1583 | No | ||
| 67 | CHN2 | 13880 | -1.301 | 0.1018 | No | ||
| 68 | BCL11A | 13924 | -1.312 | 0.1075 | No | ||
| 69 | NFIA | 14031 | -1.334 | 0.1099 | No | ||
| 70 | GRIK2 | 14212 | -1.381 | 0.1087 | No | ||
| 71 | STAT5B | 14749 | -1.521 | 0.0891 | No | ||
| 72 | ST18 | 14785 | -1.529 | 0.0965 | No | ||
| 73 | KLF12 | 15631 | -1.785 | 0.0619 | No | ||
| 74 | NEUROD1 | 16044 | -1.961 | 0.0516 | No | ||
| 75 | TACC2 | 16399 | -2.117 | 0.0455 | No | ||
| 76 | HIVEP3 | 16770 | -2.305 | 0.0396 | No | ||
| 77 | KCNA3 | 16876 | -2.365 | 0.0484 | No | ||
| 78 | PSMF1 | 16925 | -2.396 | 0.0605 | No | ||
| 79 | CAPN3 | 18510 | -5.012 | 0.0057 | No |
