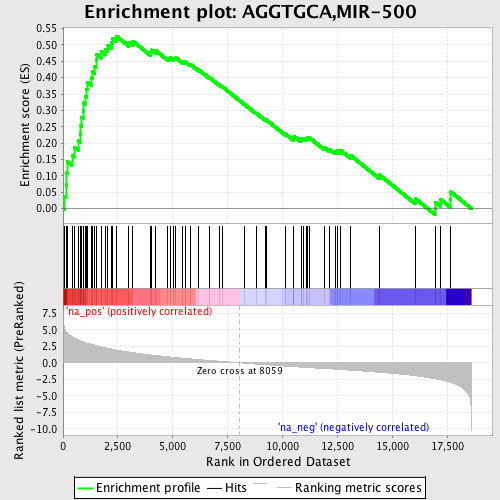
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | AGGTGCA,MIR-500 |
| Enrichment Score (ES) | 0.5267117 |
| Normalized Enrichment Score (NES) | 2.1770997 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.7373782E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CUTL1 | 73 | 5.228 | 0.0379 | Yes | ||
| 2 | RAB14 | 132 | 4.754 | 0.0728 | Yes | ||
| 3 | IVNS1ABP | 152 | 4.681 | 0.1092 | Yes | ||
| 4 | KBTBD2 | 186 | 4.581 | 0.1440 | Yes | ||
| 5 | SLC4A7 | 408 | 3.959 | 0.1638 | Yes | ||
| 6 | DCTN4 | 522 | 3.748 | 0.1877 | Yes | ||
| 7 | HIPK1 | 686 | 3.486 | 0.2068 | Yes | ||
| 8 | KLF9 | 775 | 3.344 | 0.2288 | Yes | ||
| 9 | PPP2R5E | 809 | 3.301 | 0.2534 | Yes | ||
| 10 | DDX3X | 840 | 3.265 | 0.2779 | Yes | ||
| 11 | SOCS2 | 916 | 3.168 | 0.2992 | Yes | ||
| 12 | CREB1 | 943 | 3.133 | 0.3228 | Yes | ||
| 13 | APRIN | 1027 | 3.058 | 0.3428 | Yes | ||
| 14 | TIMM8A | 1069 | 3.013 | 0.3647 | Yes | ||
| 15 | MYNN | 1112 | 2.952 | 0.3860 | Yes | ||
| 16 | FBXW7 | 1279 | 2.806 | 0.3995 | Yes | ||
| 17 | CAMK4 | 1344 | 2.749 | 0.4181 | Yes | ||
| 18 | SORCS1 | 1444 | 2.652 | 0.4339 | Yes | ||
| 19 | RBMS1 | 1499 | 2.594 | 0.4518 | Yes | ||
| 20 | PPP4R1 | 1515 | 2.586 | 0.4716 | Yes | ||
| 21 | NCOA1 | 1735 | 2.409 | 0.4791 | Yes | ||
| 22 | KPNA3 | 1922 | 2.272 | 0.4872 | Yes | ||
| 23 | E2F3 | 2042 | 2.193 | 0.4984 | Yes | ||
| 24 | PURB | 2200 | 2.082 | 0.5065 | Yes | ||
| 25 | OGT | 2242 | 2.058 | 0.5208 | Yes | ||
| 26 | UBE2D3 | 2421 | 1.941 | 0.5267 | Yes | ||
| 27 | MATR3 | 2999 | 1.615 | 0.5085 | No | ||
| 28 | NRF1 | 3183 | 1.530 | 0.5109 | No | ||
| 29 | FIGN | 4000 | 1.180 | 0.4763 | No | ||
| 30 | ABCC4 | 4024 | 1.169 | 0.4844 | No | ||
| 31 | DRD1 | 4214 | 1.102 | 0.4831 | No | ||
| 32 | NFAT5 | 4779 | 0.904 | 0.4599 | No | ||
| 33 | DDX3Y | 4895 | 0.868 | 0.4606 | No | ||
| 34 | SEC24C | 5035 | 0.824 | 0.4597 | No | ||
| 35 | RNF38 | 5143 | 0.790 | 0.4603 | No | ||
| 36 | ESRRG | 5457 | 0.691 | 0.4489 | No | ||
| 37 | CACNB1 | 5567 | 0.657 | 0.4483 | No | ||
| 38 | PRKCE | 5785 | 0.599 | 0.4414 | No | ||
| 39 | THAP11 | 6181 | 0.485 | 0.4240 | No | ||
| 40 | NOLA2 | 6658 | 0.356 | 0.4012 | No | ||
| 41 | SPG3A | 7124 | 0.231 | 0.3779 | No | ||
| 42 | RAB21 | 7273 | 0.198 | 0.3715 | No | ||
| 43 | CIT | 8259 | -0.050 | 0.3188 | No | ||
| 44 | BTBD11 | 8804 | -0.188 | 0.2910 | No | ||
| 45 | SHPRH | 9234 | -0.286 | 0.2702 | No | ||
| 46 | GABRB3 | 9250 | -0.288 | 0.2717 | No | ||
| 47 | HNRPA0 | 10123 | -0.494 | 0.2286 | No | ||
| 48 | CHRD | 10483 | -0.571 | 0.2138 | No | ||
| 49 | TBX2 | 10516 | -0.579 | 0.2167 | No | ||
| 50 | PLXDC2 | 10522 | -0.580 | 0.2211 | No | ||
| 51 | GPM6A | 10842 | -0.651 | 0.2091 | No | ||
| 52 | SLC25A26 | 10864 | -0.655 | 0.2132 | No | ||
| 53 | TSSK2 | 10958 | -0.674 | 0.2136 | No | ||
| 54 | HSPA14 | 11088 | -0.700 | 0.2122 | No | ||
| 55 | HLF | 11123 | -0.709 | 0.2161 | No | ||
| 56 | ING4 | 11208 | -0.726 | 0.2173 | No | ||
| 57 | LBP | 11902 | -0.869 | 0.1869 | No | ||
| 58 | OSBPL10 | 12154 | -0.918 | 0.1807 | No | ||
| 59 | LRRTM3 | 12392 | -0.968 | 0.1757 | No | ||
| 60 | RAI2 | 12508 | -0.990 | 0.1774 | No | ||
| 61 | PHOX2B | 12652 | -1.023 | 0.1779 | No | ||
| 62 | SYT8 | 13106 | -1.113 | 0.1624 | No | ||
| 63 | PPP3CC | 14410 | -1.434 | 0.1036 | No | ||
| 64 | CELSR3 | 16042 | -1.960 | 0.0313 | No | ||
| 65 | SP8 | 16952 | -2.417 | 0.0016 | No | ||
| 66 | POFUT1 | 16969 | -2.428 | 0.0202 | No | ||
| 67 | EGLN2 | 17197 | -2.583 | 0.0286 | No | ||
| 68 | RNF40 | 17658 | -2.982 | 0.0277 | No | ||
| 69 | RTN4RL1 | 17670 | -2.991 | 0.0510 | No |
