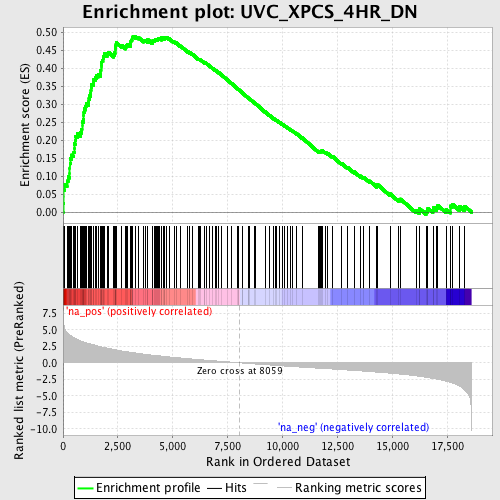
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | 0.48863834 |
| Normalized Enrichment Score (NES) | 2.3502514 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 5 | 7.263 | 0.0238 | Yes | ||
| 2 | CDYL | 20 | 6.035 | 0.0430 | Yes | ||
| 3 | APPBP2 | 21 | 5.998 | 0.0629 | Yes | ||
| 4 | CUTL1 | 73 | 5.228 | 0.0774 | Yes | ||
| 5 | AKAP10 | 180 | 4.593 | 0.0869 | Yes | ||
| 6 | CDK8 | 229 | 4.436 | 0.0990 | Yes | ||
| 7 | SEC24B | 306 | 4.204 | 0.1088 | Yes | ||
| 8 | PAFAH1B1 | 309 | 4.189 | 0.1225 | Yes | ||
| 9 | KLF7 | 313 | 4.177 | 0.1362 | Yes | ||
| 10 | SRPK2 | 340 | 4.121 | 0.1484 | Yes | ||
| 11 | MTAP | 392 | 3.994 | 0.1589 | Yes | ||
| 12 | IFRD1 | 487 | 3.805 | 0.1664 | Yes | ||
| 13 | MYST4 | 505 | 3.769 | 0.1779 | Yes | ||
| 14 | TLK1 | 510 | 3.763 | 0.1902 | Yes | ||
| 15 | CHD1 | 549 | 3.690 | 0.2003 | Yes | ||
| 16 | ATXN2 | 572 | 3.659 | 0.2113 | Yes | ||
| 17 | FCHSD2 | 651 | 3.547 | 0.2188 | Yes | ||
| 18 | ZZZ3 | 797 | 3.318 | 0.2219 | Yes | ||
| 19 | CNOT2 | 843 | 3.262 | 0.2303 | Yes | ||
| 20 | PARG | 861 | 3.235 | 0.2401 | Yes | ||
| 21 | CENTG2 | 863 | 3.229 | 0.2507 | Yes | ||
| 22 | PPP1R12A | 918 | 3.166 | 0.2583 | Yes | ||
| 23 | CREB1 | 943 | 3.133 | 0.2673 | Yes | ||
| 24 | REV3L | 948 | 3.128 | 0.2775 | Yes | ||
| 25 | ORC5L | 964 | 3.115 | 0.2870 | Yes | ||
| 26 | APRIN | 1027 | 3.058 | 0.2937 | Yes | ||
| 27 | BMPR1A | 1049 | 3.031 | 0.3026 | Yes | ||
| 28 | TLK2 | 1162 | 2.905 | 0.3062 | Yes | ||
| 29 | WDR37 | 1177 | 2.897 | 0.3150 | Yes | ||
| 30 | PTPN12 | 1209 | 2.873 | 0.3228 | Yes | ||
| 31 | BLM | 1250 | 2.831 | 0.3300 | Yes | ||
| 32 | TLE4 | 1269 | 2.814 | 0.3384 | Yes | ||
| 33 | PUM2 | 1273 | 2.810 | 0.3475 | Yes | ||
| 34 | TRIM33 | 1293 | 2.795 | 0.3558 | Yes | ||
| 35 | MARCH7 | 1377 | 2.719 | 0.3603 | Yes | ||
| 36 | CREBBP | 1398 | 2.699 | 0.3681 | Yes | ||
| 37 | EIF2C2 | 1463 | 2.637 | 0.3734 | Yes | ||
| 38 | MARK3 | 1518 | 2.584 | 0.3790 | Yes | ||
| 39 | CNOT4 | 1591 | 2.520 | 0.3834 | Yes | ||
| 40 | RNGTT | 1706 | 2.429 | 0.3853 | Yes | ||
| 41 | STARD13 | 1713 | 2.426 | 0.3930 | Yes | ||
| 42 | TIAM1 | 1727 | 2.415 | 0.4003 | Yes | ||
| 43 | ZNF292 | 1729 | 2.413 | 0.4082 | Yes | ||
| 44 | NCOA1 | 1735 | 2.409 | 0.4159 | Yes | ||
| 45 | AKAP9 | 1772 | 2.382 | 0.4219 | Yes | ||
| 46 | RBM39 | 1819 | 2.350 | 0.4272 | Yes | ||
| 47 | CTBP2 | 1847 | 2.322 | 0.4334 | Yes | ||
| 48 | RBM16 | 1874 | 2.302 | 0.4396 | Yes | ||
| 49 | MECP2 | 2004 | 2.221 | 0.4400 | Yes | ||
| 50 | RUNX1 | 2071 | 2.175 | 0.4436 | Yes | ||
| 51 | CYR61 | 2314 | 2.003 | 0.4371 | Yes | ||
| 52 | UGCG | 2342 | 1.982 | 0.4422 | Yes | ||
| 53 | ITSN2 | 2367 | 1.969 | 0.4474 | Yes | ||
| 54 | THRAP2 | 2369 | 1.968 | 0.4539 | Yes | ||
| 55 | RASA1 | 2386 | 1.958 | 0.4595 | Yes | ||
| 56 | ZFYVE9 | 2408 | 1.951 | 0.4648 | Yes | ||
| 57 | FYN | 2423 | 1.940 | 0.4705 | Yes | ||
| 58 | ANKRD17 | 2662 | 1.801 | 0.4636 | Yes | ||
| 59 | SMURF2 | 2861 | 1.683 | 0.4584 | Yes | ||
| 60 | STRN3 | 2870 | 1.679 | 0.4635 | Yes | ||
| 61 | UPF2 | 2929 | 1.648 | 0.4658 | Yes | ||
| 62 | KLF6 | 3060 | 1.589 | 0.4640 | Yes | ||
| 63 | ATF2 | 3062 | 1.588 | 0.4693 | Yes | ||
| 64 | IL6 | 3087 | 1.574 | 0.4732 | Yes | ||
| 65 | MYO10 | 3097 | 1.569 | 0.4779 | Yes | ||
| 66 | NRIP1 | 3141 | 1.549 | 0.4807 | Yes | ||
| 67 | PIP5K3 | 3142 | 1.549 | 0.4858 | Yes | ||
| 68 | CBLB | 3184 | 1.529 | 0.4886 | Yes | ||
| 69 | GLRB | 3304 | 1.476 | 0.4871 | No | ||
| 70 | NCOA3 | 3425 | 1.422 | 0.4853 | No | ||
| 71 | STK3 | 3668 | 1.312 | 0.4765 | No | ||
| 72 | THRAP1 | 3741 | 1.280 | 0.4768 | No | ||
| 73 | PTPRK | 3846 | 1.237 | 0.4753 | No | ||
| 74 | ACSL3 | 3861 | 1.230 | 0.4786 | No | ||
| 75 | MALT1 | 4076 | 1.154 | 0.4708 | No | ||
| 76 | ARFGEF1 | 4077 | 1.152 | 0.4747 | No | ||
| 77 | GNAI1 | 4092 | 1.143 | 0.4777 | No | ||
| 78 | IRAK1BP1 | 4177 | 1.116 | 0.4768 | No | ||
| 79 | DAPK1 | 4208 | 1.103 | 0.4788 | No | ||
| 80 | PVRL3 | 4277 | 1.076 | 0.4787 | No | ||
| 81 | SWAP70 | 4323 | 1.062 | 0.4798 | No | ||
| 82 | SOS2 | 4356 | 1.048 | 0.4815 | No | ||
| 83 | TCF7L2 | 4399 | 1.033 | 0.4827 | No | ||
| 84 | DYRK1A | 4492 | 1.003 | 0.4810 | No | ||
| 85 | FGF5 | 4493 | 1.003 | 0.4843 | No | ||
| 86 | STAM | 4560 | 0.977 | 0.4840 | No | ||
| 87 | BHLHB2 | 4633 | 0.952 | 0.4832 | No | ||
| 88 | MYC | 4642 | 0.949 | 0.4860 | No | ||
| 89 | TOPBP1 | 4715 | 0.926 | 0.4851 | No | ||
| 90 | BARD1 | 4829 | 0.888 | 0.4819 | No | ||
| 91 | F3 | 5054 | 0.817 | 0.4725 | No | ||
| 92 | FNDC3A | 5146 | 0.789 | 0.4702 | No | ||
| 93 | IL1RAP | 5353 | 0.726 | 0.4614 | No | ||
| 94 | ARHGAP12 | 5674 | 0.626 | 0.4461 | No | ||
| 95 | TGFBR3 | 5779 | 0.600 | 0.4425 | No | ||
| 96 | TSPAN5 | 5894 | 0.570 | 0.4382 | No | ||
| 97 | EHBP1 | 6166 | 0.487 | 0.4251 | No | ||
| 98 | BICD1 | 6197 | 0.481 | 0.4251 | No | ||
| 99 | SCHIP1 | 6279 | 0.458 | 0.4222 | No | ||
| 100 | ZNF148 | 6459 | 0.410 | 0.4139 | No | ||
| 101 | E2F5 | 6462 | 0.409 | 0.4151 | No | ||
| 102 | ACVR1 | 6525 | 0.393 | 0.4131 | No | ||
| 103 | TGFB2 | 6672 | 0.352 | 0.4063 | No | ||
| 104 | TIPARP | 6828 | 0.312 | 0.3989 | No | ||
| 105 | VEGFC | 6943 | 0.280 | 0.3937 | No | ||
| 106 | CLASP2 | 6981 | 0.268 | 0.3926 | No | ||
| 107 | USP15 | 7101 | 0.236 | 0.3869 | No | ||
| 108 | ARHGEF7 | 7199 | 0.214 | 0.3824 | No | ||
| 109 | IGF1R | 7496 | 0.141 | 0.3668 | No | ||
| 110 | PLEKHC1 | 7659 | 0.100 | 0.3583 | No | ||
| 111 | SHB | 7925 | 0.033 | 0.3441 | No | ||
| 112 | PCTK2 | 7984 | 0.019 | 0.3410 | No | ||
| 113 | KLF10 | 8004 | 0.013 | 0.3400 | No | ||
| 114 | MTF2 | 8155 | -0.021 | 0.3319 | No | ||
| 115 | CTGF | 8162 | -0.024 | 0.3317 | No | ||
| 116 | NUP153 | 8465 | -0.107 | 0.3157 | No | ||
| 117 | RRAS2 | 8468 | -0.107 | 0.3159 | No | ||
| 118 | RCOR1 | 8494 | -0.114 | 0.3149 | No | ||
| 119 | FBXW11 | 8723 | -0.167 | 0.3031 | No | ||
| 120 | CENTD1 | 8756 | -0.176 | 0.3020 | No | ||
| 121 | SAMD4A | 9203 | -0.280 | 0.2787 | No | ||
| 122 | SATB2 | 9416 | -0.321 | 0.2683 | No | ||
| 123 | EXT1 | 9599 | -0.367 | 0.2597 | No | ||
| 124 | CENPA | 9683 | -0.387 | 0.2564 | No | ||
| 125 | RGS7 | 9727 | -0.396 | 0.2554 | No | ||
| 126 | MICAL3 | 9849 | -0.426 | 0.2503 | No | ||
| 127 | NFIB | 9981 | -0.460 | 0.2447 | No | ||
| 128 | TAF4 | 10103 | -0.489 | 0.2398 | No | ||
| 129 | ZHX2 | 10220 | -0.512 | 0.2352 | No | ||
| 130 | PHLPP | 10366 | -0.545 | 0.2291 | No | ||
| 131 | CENPE | 10476 | -0.570 | 0.2251 | No | ||
| 132 | SPEN | 10640 | -0.606 | 0.2183 | No | ||
| 133 | TUBGCP3 | 10891 | -0.662 | 0.2069 | No | ||
| 134 | SOS1 | 11653 | -0.819 | 0.1684 | No | ||
| 135 | DNMBP | 11680 | -0.822 | 0.1697 | No | ||
| 136 | ANKS1A | 11737 | -0.834 | 0.1694 | No | ||
| 137 | MID1 | 11775 | -0.841 | 0.1702 | No | ||
| 138 | KIF2A | 11815 | -0.851 | 0.1709 | No | ||
| 139 | ZCCHC11 | 11941 | -0.877 | 0.1670 | No | ||
| 140 | EPHA4 | 12057 | -0.898 | 0.1638 | No | ||
| 141 | FBXL7 | 12281 | -0.945 | 0.1548 | No | ||
| 142 | CD2AP | 12709 | -1.034 | 0.1351 | No | ||
| 143 | RNF13 | 12954 | -1.085 | 0.1254 | No | ||
| 144 | ZHX3 | 13270 | -1.152 | 0.1122 | No | ||
| 145 | HOXB2 | 13556 | -1.219 | 0.1008 | No | ||
| 146 | BCAR3 | 13691 | -1.256 | 0.0977 | No | ||
| 147 | PBX3 | 13972 | -1.320 | 0.0869 | No | ||
| 148 | WNT5A | 14294 | -1.403 | 0.0741 | No | ||
| 149 | DDX10 | 14338 | -1.415 | 0.0765 | No | ||
| 150 | CENPC1 | 14902 | -1.563 | 0.0511 | No | ||
| 151 | TERF1 | 15278 | -1.677 | 0.0364 | No | ||
| 152 | MTMR3 | 15375 | -1.708 | 0.0368 | No | ||
| 153 | SOCS5 | 16084 | -1.974 | 0.0050 | No | ||
| 154 | MYST3 | 16231 | -2.036 | 0.0038 | No | ||
| 155 | NCK1 | 16253 | -2.044 | 0.0094 | No | ||
| 156 | LHFPL2 | 16542 | -2.181 | 0.0010 | No | ||
| 157 | SRGAP2 | 16591 | -2.210 | 0.0057 | No | ||
| 158 | SLC25A12 | 16625 | -2.225 | 0.0113 | No | ||
| 159 | BTRC | 16859 | -2.357 | 0.0065 | No | ||
| 160 | TLE1 | 16890 | -2.374 | 0.0127 | No | ||
| 161 | DOCK4 | 17027 | -2.465 | 0.0135 | No | ||
| 162 | FGF2 | 17079 | -2.496 | 0.0190 | No | ||
| 163 | EGFR | 17460 | -2.782 | 0.0076 | No | ||
| 164 | GNAQ | 17642 | -2.964 | 0.0076 | No | ||
| 165 | HIRA | 17643 | -2.964 | 0.0174 | No | ||
| 166 | PRR3 | 17730 | -3.053 | 0.0229 | No | ||
| 167 | DLC1 | 18054 | -3.505 | 0.0170 | No | ||
| 168 | TRIM2 | 18296 | -4.052 | 0.0173 | No |
