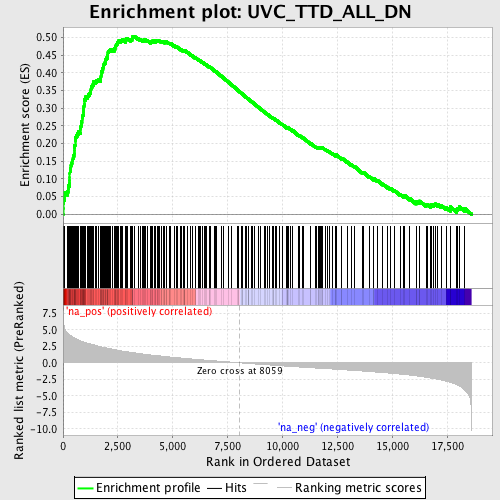
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | UVC_TTD_ALL_DN |
| Enrichment Score (ES) | 0.5033847 |
| Normalized Enrichment Score (NES) | 2.5246434 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 5 | 7.263 | 0.0154 | Yes | ||
| 2 | SSBP2 | 9 | 6.794 | 0.0299 | Yes | ||
| 3 | CDYL | 20 | 6.035 | 0.0424 | Yes | ||
| 4 | CUTL1 | 73 | 5.228 | 0.0508 | Yes | ||
| 5 | FUBP1 | 83 | 5.091 | 0.0613 | Yes | ||
| 6 | EIF4G3 | 211 | 4.479 | 0.0640 | Yes | ||
| 7 | CDK8 | 229 | 4.436 | 0.0727 | Yes | ||
| 8 | PRKACB | 246 | 4.348 | 0.0812 | Yes | ||
| 9 | MAPK6 | 289 | 4.244 | 0.0880 | Yes | ||
| 10 | SLC25A24 | 300 | 4.226 | 0.0966 | Yes | ||
| 11 | SEC24B | 306 | 4.204 | 0.1054 | Yes | ||
| 12 | PAFAH1B1 | 309 | 4.189 | 0.1143 | Yes | ||
| 13 | KLF7 | 313 | 4.177 | 0.1232 | Yes | ||
| 14 | SRPK2 | 340 | 4.121 | 0.1307 | Yes | ||
| 15 | HNRPDL | 357 | 4.077 | 0.1386 | Yes | ||
| 16 | DHX15 | 394 | 3.982 | 0.1452 | Yes | ||
| 17 | SLC4A7 | 408 | 3.959 | 0.1530 | Yes | ||
| 18 | SOCS6 | 449 | 3.868 | 0.1592 | Yes | ||
| 19 | IFRD1 | 487 | 3.805 | 0.1654 | Yes | ||
| 20 | UBL3 | 499 | 3.780 | 0.1729 | Yes | ||
| 21 | MYST4 | 505 | 3.769 | 0.1808 | Yes | ||
| 22 | TLK1 | 510 | 3.763 | 0.1887 | Yes | ||
| 23 | SPOP | 529 | 3.732 | 0.1958 | Yes | ||
| 24 | MNAT1 | 548 | 3.691 | 0.2027 | Yes | ||
| 25 | CHD1 | 549 | 3.690 | 0.2107 | Yes | ||
| 26 | ATXN2 | 572 | 3.659 | 0.2174 | Yes | ||
| 27 | MAP2K4 | 603 | 3.616 | 0.2236 | Yes | ||
| 28 | FCHSD2 | 651 | 3.547 | 0.2287 | Yes | ||
| 29 | MPP6 | 692 | 3.473 | 0.2340 | Yes | ||
| 30 | MBNL1 | 795 | 3.320 | 0.2356 | Yes | ||
| 31 | ZZZ3 | 797 | 3.318 | 0.2427 | Yes | ||
| 32 | PPP2R5E | 809 | 3.301 | 0.2492 | Yes | ||
| 33 | SLC25A13 | 821 | 3.281 | 0.2557 | Yes | ||
| 34 | CNOT2 | 843 | 3.262 | 0.2616 | Yes | ||
| 35 | PARG | 861 | 3.235 | 0.2676 | Yes | ||
| 36 | CENTG2 | 863 | 3.229 | 0.2745 | Yes | ||
| 37 | SIPA1L1 | 900 | 3.187 | 0.2794 | Yes | ||
| 38 | PPP1R12A | 918 | 3.166 | 0.2853 | Yes | ||
| 39 | UST | 921 | 3.165 | 0.2921 | Yes | ||
| 40 | PKN2 | 941 | 3.137 | 0.2978 | Yes | ||
| 41 | REV3L | 948 | 3.128 | 0.3042 | Yes | ||
| 42 | LRP6 | 955 | 3.124 | 0.3106 | Yes | ||
| 43 | ORC5L | 964 | 3.115 | 0.3169 | Yes | ||
| 44 | ZMYND11 | 965 | 3.111 | 0.3236 | Yes | ||
| 45 | ABI1 | 1014 | 3.065 | 0.3276 | Yes | ||
| 46 | GSK3B | 1038 | 3.046 | 0.3329 | Yes | ||
| 47 | ATBF1 | 1132 | 2.935 | 0.3342 | Yes | ||
| 48 | TLK2 | 1162 | 2.905 | 0.3389 | Yes | ||
| 49 | PTPN12 | 1209 | 2.873 | 0.3426 | Yes | ||
| 50 | BLM | 1250 | 2.831 | 0.3465 | Yes | ||
| 51 | TLE4 | 1269 | 2.814 | 0.3516 | Yes | ||
| 52 | PUM2 | 1273 | 2.810 | 0.3575 | Yes | ||
| 53 | TRIM33 | 1293 | 2.795 | 0.3625 | Yes | ||
| 54 | CLASP1 | 1347 | 2.744 | 0.3655 | Yes | ||
| 55 | MARCH7 | 1377 | 2.719 | 0.3698 | Yes | ||
| 56 | CREBBP | 1398 | 2.699 | 0.3745 | Yes | ||
| 57 | LASS6 | 1476 | 2.625 | 0.3760 | Yes | ||
| 58 | TBL1X | 1534 | 2.569 | 0.3784 | Yes | ||
| 59 | CNOT4 | 1591 | 2.520 | 0.3808 | Yes | ||
| 60 | RNGTT | 1706 | 2.429 | 0.3798 | Yes | ||
| 61 | STARD13 | 1713 | 2.426 | 0.3847 | Yes | ||
| 62 | ARIH1 | 1724 | 2.416 | 0.3894 | Yes | ||
| 63 | ZNF292 | 1729 | 2.413 | 0.3944 | Yes | ||
| 64 | NCOA1 | 1735 | 2.409 | 0.3993 | Yes | ||
| 65 | CTAGE5 | 1737 | 2.407 | 0.4045 | Yes | ||
| 66 | EIF4E | 1784 | 2.371 | 0.4071 | Yes | ||
| 67 | TCF12 | 1786 | 2.371 | 0.4121 | Yes | ||
| 68 | RBM39 | 1819 | 2.350 | 0.4154 | Yes | ||
| 69 | CTBP2 | 1847 | 2.322 | 0.4190 | Yes | ||
| 70 | ZRF1 | 1857 | 2.318 | 0.4235 | Yes | ||
| 71 | RBM16 | 1874 | 2.302 | 0.4276 | Yes | ||
| 72 | WDHD1 | 1926 | 2.269 | 0.4297 | Yes | ||
| 73 | XRCC4 | 1930 | 2.266 | 0.4344 | Yes | ||
| 74 | KIF23 | 1941 | 2.259 | 0.4388 | Yes | ||
| 75 | ROCK2 | 1973 | 2.242 | 0.4419 | Yes | ||
| 76 | SEMA3C | 2011 | 2.215 | 0.4447 | Yes | ||
| 77 | MDN1 | 2019 | 2.209 | 0.4490 | Yes | ||
| 78 | E2F3 | 2042 | 2.193 | 0.4526 | Yes | ||
| 79 | DOK5 | 2043 | 2.191 | 0.4573 | Yes | ||
| 80 | RUNX1 | 2071 | 2.175 | 0.4605 | Yes | ||
| 81 | WDFY3 | 2108 | 2.150 | 0.4632 | Yes | ||
| 82 | STAG1 | 2159 | 2.109 | 0.4650 | Yes | ||
| 83 | OGT | 2242 | 2.058 | 0.4650 | Yes | ||
| 84 | INPP5F | 2324 | 1.998 | 0.4649 | Yes | ||
| 85 | UGCG | 2342 | 1.982 | 0.4682 | Yes | ||
| 86 | ITSN2 | 2367 | 1.969 | 0.4712 | Yes | ||
| 87 | THRAP2 | 2369 | 1.968 | 0.4754 | Yes | ||
| 88 | ZFYVE9 | 2408 | 1.951 | 0.4775 | Yes | ||
| 89 | FYN | 2423 | 1.940 | 0.4809 | Yes | ||
| 90 | WDR7 | 2465 | 1.915 | 0.4828 | Yes | ||
| 91 | RBM6 | 2479 | 1.907 | 0.4862 | Yes | ||
| 92 | ENC1 | 2513 | 1.886 | 0.4885 | Yes | ||
| 93 | PPAP2B | 2543 | 1.868 | 0.4910 | Yes | ||
| 94 | MSH3 | 2634 | 1.816 | 0.4900 | Yes | ||
| 95 | ANKRD17 | 2662 | 1.801 | 0.4924 | Yes | ||
| 96 | PKP4 | 2708 | 1.767 | 0.4937 | Yes | ||
| 97 | SMURF2 | 2861 | 1.683 | 0.4891 | Yes | ||
| 98 | AQR | 2864 | 1.682 | 0.4926 | Yes | ||
| 99 | STRN3 | 2870 | 1.679 | 0.4960 | Yes | ||
| 100 | UPF2 | 2929 | 1.648 | 0.4964 | Yes | ||
| 101 | ATF2 | 3062 | 1.588 | 0.4926 | Yes | ||
| 102 | MYO10 | 3097 | 1.569 | 0.4941 | Yes | ||
| 103 | NRIP1 | 3141 | 1.549 | 0.4951 | Yes | ||
| 104 | PIP5K3 | 3142 | 1.549 | 0.4985 | Yes | ||
| 105 | EVI5 | 3154 | 1.543 | 0.5012 | Yes | ||
| 106 | CBLB | 3184 | 1.529 | 0.5029 | Yes | ||
| 107 | ZNF638 | 3236 | 1.511 | 0.5034 | Yes | ||
| 108 | NCOA3 | 3425 | 1.422 | 0.4962 | No | ||
| 109 | CASK | 3512 | 1.379 | 0.4945 | No | ||
| 110 | SPAG9 | 3636 | 1.325 | 0.4907 | No | ||
| 111 | STK3 | 3668 | 1.312 | 0.4918 | No | ||
| 112 | MELK | 3695 | 1.301 | 0.4932 | No | ||
| 113 | THRAP1 | 3741 | 1.280 | 0.4935 | No | ||
| 114 | PTPRK | 3846 | 1.237 | 0.4905 | No | ||
| 115 | RGS20 | 3991 | 1.183 | 0.4852 | No | ||
| 116 | FUT8 | 3994 | 1.181 | 0.4876 | No | ||
| 117 | PRIM2A | 4013 | 1.173 | 0.4892 | No | ||
| 118 | ARFGEF1 | 4077 | 1.152 | 0.4882 | No | ||
| 119 | GNAI1 | 4092 | 1.143 | 0.4899 | No | ||
| 120 | IRAK1BP1 | 4177 | 1.116 | 0.4878 | No | ||
| 121 | DAPK1 | 4208 | 1.103 | 0.4885 | No | ||
| 122 | CCND1 | 4211 | 1.103 | 0.4908 | No | ||
| 123 | LRIG1 | 4281 | 1.076 | 0.4894 | No | ||
| 124 | SWAP70 | 4323 | 1.062 | 0.4894 | No | ||
| 125 | CDC42BPA | 4353 | 1.049 | 0.4901 | No | ||
| 126 | TCF7L2 | 4399 | 1.033 | 0.4899 | No | ||
| 127 | DYRK1A | 4492 | 1.003 | 0.4870 | No | ||
| 128 | FGF5 | 4493 | 1.003 | 0.4892 | No | ||
| 129 | TDRD3 | 4583 | 0.968 | 0.4864 | No | ||
| 130 | BHLHB2 | 4633 | 0.952 | 0.4858 | No | ||
| 131 | MYC | 4642 | 0.949 | 0.4874 | No | ||
| 132 | COL5A2 | 4707 | 0.928 | 0.4859 | No | ||
| 133 | TOPBP1 | 4715 | 0.926 | 0.4875 | No | ||
| 134 | BARD1 | 4829 | 0.888 | 0.4833 | No | ||
| 135 | MPHOSPH1 | 4861 | 0.880 | 0.4835 | No | ||
| 136 | CASP8 | 4914 | 0.862 | 0.4825 | No | ||
| 137 | F3 | 5054 | 0.817 | 0.4767 | No | ||
| 138 | FNDC3A | 5146 | 0.789 | 0.4735 | No | ||
| 139 | MITF | 5192 | 0.775 | 0.4727 | No | ||
| 140 | IL1RAP | 5353 | 0.726 | 0.4655 | No | ||
| 141 | STK39 | 5404 | 0.711 | 0.4644 | No | ||
| 142 | JMJD2C | 5500 | 0.680 | 0.4606 | No | ||
| 143 | LARS2 | 5510 | 0.677 | 0.4616 | No | ||
| 144 | MGLL | 5522 | 0.673 | 0.4625 | No | ||
| 145 | DPYD | 5538 | 0.668 | 0.4631 | No | ||
| 146 | ARHGAP12 | 5674 | 0.626 | 0.4571 | No | ||
| 147 | MAST2 | 5816 | 0.590 | 0.4507 | No | ||
| 148 | TSPAN5 | 5894 | 0.570 | 0.4477 | No | ||
| 149 | SLC16A7 | 6011 | 0.533 | 0.4425 | No | ||
| 150 | GOLGA4 | 6013 | 0.532 | 0.4436 | No | ||
| 151 | EHBP1 | 6166 | 0.487 | 0.4364 | No | ||
| 152 | BICD1 | 6197 | 0.481 | 0.4358 | No | ||
| 153 | SCHIP1 | 6279 | 0.458 | 0.4324 | No | ||
| 154 | FLNB | 6345 | 0.441 | 0.4298 | No | ||
| 155 | ZNF148 | 6459 | 0.410 | 0.4245 | No | ||
| 156 | SERPINB2 | 6491 | 0.402 | 0.4237 | No | ||
| 157 | ACVR1 | 6525 | 0.393 | 0.4227 | No | ||
| 158 | MAD1L1 | 6651 | 0.358 | 0.4167 | No | ||
| 159 | TGFB2 | 6672 | 0.352 | 0.4164 | No | ||
| 160 | AUH | 6696 | 0.346 | 0.4159 | No | ||
| 161 | GTF2F2 | 6887 | 0.294 | 0.4062 | No | ||
| 162 | VEGFC | 6943 | 0.280 | 0.4038 | No | ||
| 163 | CLASP2 | 6981 | 0.268 | 0.4023 | No | ||
| 164 | ARHGEF7 | 7199 | 0.214 | 0.3910 | No | ||
| 165 | EDG2 | 7313 | 0.188 | 0.3852 | No | ||
| 166 | CRY1 | 7537 | 0.131 | 0.3733 | No | ||
| 167 | PLEKHC1 | 7659 | 0.100 | 0.3670 | No | ||
| 168 | SHB | 7925 | 0.033 | 0.3526 | No | ||
| 169 | PCTK2 | 7984 | 0.019 | 0.3495 | No | ||
| 170 | KLF10 | 8004 | 0.013 | 0.3485 | No | ||
| 171 | NEK7 | 8141 | -0.017 | 0.3411 | No | ||
| 172 | MTF2 | 8155 | -0.021 | 0.3404 | No | ||
| 173 | CTGF | 8162 | -0.024 | 0.3402 | No | ||
| 174 | ARNT2 | 8292 | -0.058 | 0.3333 | No | ||
| 175 | TRIM24 | 8354 | -0.075 | 0.3301 | No | ||
| 176 | DUSP1 | 8464 | -0.107 | 0.3244 | No | ||
| 177 | NUP153 | 8465 | -0.107 | 0.3246 | No | ||
| 178 | RRAS2 | 8468 | -0.107 | 0.3247 | No | ||
| 179 | FTO | 8605 | -0.141 | 0.3176 | No | ||
| 180 | RGL1 | 8623 | -0.144 | 0.3170 | No | ||
| 181 | FBXW11 | 8723 | -0.167 | 0.3120 | No | ||
| 182 | FARS2 | 8906 | -0.210 | 0.3025 | No | ||
| 183 | RABGAP1L | 8976 | -0.225 | 0.2993 | No | ||
| 184 | ITSN1 | 9186 | -0.277 | 0.2885 | No | ||
| 185 | SAMD4A | 9203 | -0.280 | 0.2882 | No | ||
| 186 | LYN | 9318 | -0.303 | 0.2826 | No | ||
| 187 | SATB2 | 9416 | -0.321 | 0.2780 | No | ||
| 188 | MYO1B | 9537 | -0.352 | 0.2723 | No | ||
| 189 | NRG1 | 9589 | -0.363 | 0.2703 | No | ||
| 190 | EXT1 | 9599 | -0.367 | 0.2706 | No | ||
| 191 | CENPA | 9683 | -0.387 | 0.2669 | No | ||
| 192 | RGS7 | 9727 | -0.396 | 0.2654 | No | ||
| 193 | CRADD | 9868 | -0.432 | 0.2587 | No | ||
| 194 | NFIB | 9981 | -0.460 | 0.2536 | No | ||
| 195 | ATXN1 | 9983 | -0.460 | 0.2545 | No | ||
| 196 | PTPRM | 10196 | -0.508 | 0.2441 | No | ||
| 197 | ZHX2 | 10220 | -0.512 | 0.2439 | No | ||
| 198 | GBE1 | 10242 | -0.517 | 0.2439 | No | ||
| 199 | IL7R | 10247 | -0.518 | 0.2448 | No | ||
| 200 | ROR1 | 10279 | -0.525 | 0.2442 | No | ||
| 201 | PHLPP | 10366 | -0.545 | 0.2407 | No | ||
| 202 | CENPE | 10476 | -0.570 | 0.2360 | No | ||
| 203 | SEPT9 | 10734 | -0.626 | 0.2234 | No | ||
| 204 | SMAD7 | 10786 | -0.638 | 0.2220 | No | ||
| 205 | TUBGCP3 | 10891 | -0.662 | 0.2177 | No | ||
| 206 | SLIT1 | 10951 | -0.672 | 0.2160 | No | ||
| 207 | SKAP2 | 11276 | -0.741 | 0.1999 | No | ||
| 208 | SYNJ2 | 11291 | -0.744 | 0.2008 | No | ||
| 209 | CXCL12 | 11524 | -0.791 | 0.1898 | No | ||
| 210 | ERC1 | 11563 | -0.799 | 0.1895 | No | ||
| 211 | TRAM2 | 11639 | -0.816 | 0.1871 | No | ||
| 212 | SOS1 | 11653 | -0.819 | 0.1882 | No | ||
| 213 | DNMBP | 11680 | -0.822 | 0.1886 | No | ||
| 214 | ANKS1A | 11737 | -0.834 | 0.1873 | No | ||
| 215 | MID1 | 11775 | -0.841 | 0.1871 | No | ||
| 216 | ACVR2A | 11808 | -0.850 | 0.1872 | No | ||
| 217 | KIF2A | 11815 | -0.851 | 0.1887 | No | ||
| 218 | ZCCHC11 | 11941 | -0.877 | 0.1838 | No | ||
| 219 | EPHA4 | 12057 | -0.898 | 0.1795 | No | ||
| 220 | ARID5B | 12148 | -0.915 | 0.1765 | No | ||
| 221 | FBXL7 | 12281 | -0.945 | 0.1714 | No | ||
| 222 | GALK2 | 12409 | -0.970 | 0.1665 | No | ||
| 223 | PTPN2 | 12427 | -0.974 | 0.1677 | No | ||
| 224 | PRKCA | 12452 | -0.980 | 0.1685 | No | ||
| 225 | BMP2K | 12688 | -1.030 | 0.1579 | No | ||
| 226 | CD2AP | 12709 | -1.034 | 0.1591 | No | ||
| 227 | RNF13 | 12954 | -1.085 | 0.1481 | No | ||
| 228 | CAP2 | 13141 | -1.120 | 0.1404 | No | ||
| 229 | ZHX3 | 13270 | -1.152 | 0.1359 | No | ||
| 230 | ATRX | 13665 | -1.248 | 0.1172 | No | ||
| 231 | BCAR3 | 13691 | -1.256 | 0.1185 | No | ||
| 232 | PBX3 | 13972 | -1.320 | 0.1061 | No | ||
| 233 | ANAPC10 | 14146 | -1.364 | 0.0996 | No | ||
| 234 | SMAD3 | 14165 | -1.369 | 0.1016 | No | ||
| 235 | DDX10 | 14338 | -1.415 | 0.0953 | No | ||
| 236 | SERPINE1 | 14567 | -1.475 | 0.0860 | No | ||
| 237 | MGAT5 | 14787 | -1.530 | 0.0774 | No | ||
| 238 | PARD3 | 14925 | -1.571 | 0.0733 | No | ||
| 239 | HOMER1 | 15100 | -1.623 | 0.0673 | No | ||
| 240 | MTMR3 | 15375 | -1.708 | 0.0561 | No | ||
| 241 | GULP1 | 15516 | -1.749 | 0.0522 | No | ||
| 242 | NUP98 | 15574 | -1.767 | 0.0529 | No | ||
| 243 | GPR176 | 15806 | -1.859 | 0.0444 | No | ||
| 244 | SOCS5 | 16084 | -1.974 | 0.0335 | No | ||
| 245 | PPP2R2A | 16113 | -1.987 | 0.0363 | No | ||
| 246 | MYST3 | 16231 | -2.036 | 0.0343 | No | ||
| 247 | MAP4K5 | 16248 | -2.043 | 0.0378 | No | ||
| 248 | LHFPL2 | 16542 | -2.181 | 0.0266 | No | ||
| 249 | SRGAP2 | 16591 | -2.210 | 0.0287 | No | ||
| 250 | CREB5 | 16764 | -2.299 | 0.0243 | No | ||
| 251 | HAS2 | 16772 | -2.306 | 0.0289 | No | ||
| 252 | TLE1 | 16890 | -2.374 | 0.0277 | No | ||
| 253 | PEX14 | 16949 | -2.415 | 0.0297 | No | ||
| 254 | FGF2 | 17079 | -2.496 | 0.0281 | No | ||
| 255 | ORC2L | 17227 | -2.606 | 0.0257 | No | ||
| 256 | EGFR | 17460 | -2.782 | 0.0190 | No | ||
| 257 | GNAQ | 17642 | -2.964 | 0.0156 | No | ||
| 258 | HIRA | 17643 | -2.964 | 0.0220 | No | ||
| 259 | DAAM1 | 17943 | -3.325 | 0.0129 | No | ||
| 260 | IGF2BP3 | 17985 | -3.386 | 0.0179 | No | ||
| 261 | DLC1 | 18054 | -3.505 | 0.0218 | No | ||
| 262 | AMPH | 18303 | -4.074 | 0.0171 | No |
