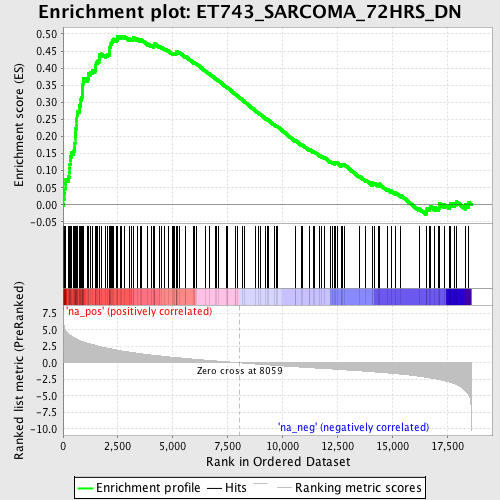
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ET743_SARCOMA_72HRS_DN |
| Enrichment Score (ES) | 0.49375042 |
| Normalized Enrichment Score (NES) | 2.3758361 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDX20 | 17 | 6.114 | 0.0180 | Yes | ||
| 2 | LBR | 43 | 5.600 | 0.0339 | Yes | ||
| 3 | SLC4A1AP | 84 | 5.078 | 0.0475 | Yes | ||
| 4 | EMP1 | 107 | 4.902 | 0.0614 | Yes | ||
| 5 | TMPO | 124 | 4.809 | 0.0754 | Yes | ||
| 6 | SMAD4 | 241 | 4.376 | 0.0827 | Yes | ||
| 7 | HSPH1 | 274 | 4.270 | 0.0941 | Yes | ||
| 8 | TXNRD1 | 288 | 4.246 | 0.1065 | Yes | ||
| 9 | HDAC2 | 308 | 4.192 | 0.1185 | Yes | ||
| 10 | ATP2A2 | 317 | 4.171 | 0.1309 | Yes | ||
| 11 | SRPK2 | 340 | 4.121 | 0.1425 | Yes | ||
| 12 | SLC25A17 | 391 | 3.994 | 0.1521 | Yes | ||
| 13 | GTF2H2 | 494 | 3.791 | 0.1583 | Yes | ||
| 14 | MAT2A | 518 | 3.752 | 0.1686 | Yes | ||
| 15 | TBK1 | 530 | 3.732 | 0.1796 | Yes | ||
| 16 | ATG5 | 544 | 3.702 | 0.1903 | Yes | ||
| 17 | RAP1B | 546 | 3.696 | 0.2017 | Yes | ||
| 18 | RAB1A | 560 | 3.680 | 0.2124 | Yes | ||
| 19 | ZDHHC5 | 574 | 3.656 | 0.2230 | Yes | ||
| 20 | PRKD3 | 596 | 3.625 | 0.2330 | Yes | ||
| 21 | MRPL1 | 600 | 3.620 | 0.2440 | Yes | ||
| 22 | HSPA8 | 629 | 3.581 | 0.2536 | Yes | ||
| 23 | GTF2H1 | 632 | 3.579 | 0.2645 | Yes | ||
| 24 | PSMA4 | 668 | 3.508 | 0.2735 | Yes | ||
| 25 | PIK3C3 | 731 | 3.412 | 0.2807 | Yes | ||
| 26 | PCNA | 752 | 3.379 | 0.2900 | Yes | ||
| 27 | RABGAP1 | 799 | 3.317 | 0.2978 | Yes | ||
| 28 | PPP2R5E | 809 | 3.301 | 0.3075 | Yes | ||
| 29 | DNAJA2 | 854 | 3.245 | 0.3152 | Yes | ||
| 30 | POLD3 | 865 | 3.229 | 0.3246 | Yes | ||
| 31 | CCT4 | 880 | 3.212 | 0.3338 | Yes | ||
| 32 | ANLN | 887 | 3.202 | 0.3433 | Yes | ||
| 33 | DDX21 | 902 | 3.184 | 0.3524 | Yes | ||
| 34 | CCNH | 910 | 3.175 | 0.3619 | Yes | ||
| 35 | PPP2CB | 936 | 3.148 | 0.3702 | Yes | ||
| 36 | ACTL6A | 1125 | 2.939 | 0.3691 | Yes | ||
| 37 | RABGGTB | 1151 | 2.918 | 0.3768 | Yes | ||
| 38 | NCOA6 | 1153 | 2.917 | 0.3858 | Yes | ||
| 39 | PSMD12 | 1264 | 2.817 | 0.3885 | Yes | ||
| 40 | SIRT1 | 1352 | 2.740 | 0.3923 | Yes | ||
| 41 | MAP3K7 | 1454 | 2.642 | 0.3949 | Yes | ||
| 42 | BIRC2 | 1468 | 2.635 | 0.4024 | Yes | ||
| 43 | CDC5L | 1491 | 2.601 | 0.4092 | Yes | ||
| 44 | EXOC2 | 1498 | 2.594 | 0.4169 | Yes | ||
| 45 | EIF3S10 | 1569 | 2.537 | 0.4210 | Yes | ||
| 46 | EIF4B | 1637 | 2.481 | 0.4250 | Yes | ||
| 47 | TCERG1 | 1651 | 2.470 | 0.4319 | Yes | ||
| 48 | TOP1 | 1664 | 2.462 | 0.4389 | Yes | ||
| 49 | CDC23 | 1753 | 2.396 | 0.4415 | Yes | ||
| 50 | WEE1 | 1943 | 2.258 | 0.4383 | Yes | ||
| 51 | FNTA | 2028 | 2.202 | 0.4405 | Yes | ||
| 52 | USP12 | 2105 | 2.153 | 0.4431 | Yes | ||
| 53 | MAPK14 | 2121 | 2.137 | 0.4488 | Yes | ||
| 54 | TOP2A | 2126 | 2.130 | 0.4552 | Yes | ||
| 55 | CCNA2 | 2128 | 2.129 | 0.4617 | Yes | ||
| 56 | UBE2D2 | 2174 | 2.100 | 0.4658 | Yes | ||
| 57 | ELOVL6 | 2179 | 2.096 | 0.4721 | Yes | ||
| 58 | GTF2B | 2218 | 2.072 | 0.4764 | Yes | ||
| 59 | OGT | 2242 | 2.058 | 0.4815 | Yes | ||
| 60 | RIPK1 | 2288 | 2.022 | 0.4853 | Yes | ||
| 61 | UBE2D3 | 2421 | 1.941 | 0.4842 | Yes | ||
| 62 | RPS7 | 2468 | 1.914 | 0.4876 | Yes | ||
| 63 | SUPV3L1 | 2481 | 1.906 | 0.4928 | Yes | ||
| 64 | SNRPG | 2614 | 1.836 | 0.4914 | Yes | ||
| 65 | MCTS1 | 2673 | 1.792 | 0.4938 | Yes | ||
| 66 | PSMC6 | 2803 | 1.716 | 0.4921 | No | ||
| 67 | MRPL15 | 3008 | 1.613 | 0.4860 | No | ||
| 68 | SON | 3127 | 1.555 | 0.4844 | No | ||
| 69 | GNL2 | 3185 | 1.529 | 0.4860 | No | ||
| 70 | SIRPA | 3195 | 1.525 | 0.4903 | No | ||
| 71 | USP3 | 3367 | 1.449 | 0.4855 | No | ||
| 72 | HSPA1L | 3521 | 1.376 | 0.4814 | No | ||
| 73 | PJA2 | 3570 | 1.359 | 0.4830 | No | ||
| 74 | ACSL3 | 3861 | 1.230 | 0.4711 | No | ||
| 75 | PRIM2A | 4013 | 1.173 | 0.4666 | No | ||
| 76 | RBM17 | 4109 | 1.140 | 0.4649 | No | ||
| 77 | RNF14 | 4143 | 1.126 | 0.4666 | No | ||
| 78 | GDI2 | 4179 | 1.115 | 0.4682 | No | ||
| 79 | PPP1CC | 4183 | 1.114 | 0.4715 | No | ||
| 80 | TNFSF5IP1 | 4397 | 1.034 | 0.4631 | No | ||
| 81 | MRPL35 | 4482 | 1.007 | 0.4617 | No | ||
| 82 | MYC | 4642 | 0.949 | 0.4560 | No | ||
| 83 | CFLAR | 4783 | 0.903 | 0.4512 | No | ||
| 84 | XBP1 | 4976 | 0.842 | 0.4434 | No | ||
| 85 | FBXL3 | 5013 | 0.828 | 0.4440 | No | ||
| 86 | GOPC | 5074 | 0.811 | 0.4432 | No | ||
| 87 | FNDC3A | 5146 | 0.789 | 0.4418 | No | ||
| 88 | CLCN7 | 5147 | 0.789 | 0.4443 | No | ||
| 89 | FAS | 5156 | 0.786 | 0.4463 | No | ||
| 90 | MCM3 | 5174 | 0.780 | 0.4478 | No | ||
| 91 | TARDBP | 5221 | 0.768 | 0.4476 | No | ||
| 92 | GGPS1 | 5315 | 0.741 | 0.4449 | No | ||
| 93 | STK17B | 5555 | 0.661 | 0.4340 | No | ||
| 94 | SSFA2 | 5597 | 0.647 | 0.4338 | No | ||
| 95 | BCL2L1 | 5956 | 0.550 | 0.4160 | No | ||
| 96 | GABPB2 | 5995 | 0.539 | 0.4157 | No | ||
| 97 | EML4 | 6090 | 0.510 | 0.4121 | No | ||
| 98 | ARFGEF2 | 6503 | 0.399 | 0.3910 | No | ||
| 99 | SPRY2 | 6663 | 0.354 | 0.3835 | No | ||
| 100 | VEGFC | 6943 | 0.280 | 0.3693 | No | ||
| 101 | RAB5A | 6995 | 0.265 | 0.3673 | No | ||
| 102 | MFAP1 | 7059 | 0.248 | 0.3647 | No | ||
| 103 | PSMB1 | 7457 | 0.152 | 0.3436 | No | ||
| 104 | SEH1L | 7473 | 0.147 | 0.3433 | No | ||
| 105 | EPS15 | 7475 | 0.146 | 0.3437 | No | ||
| 106 | TIAM2 | 7857 | 0.053 | 0.3232 | No | ||
| 107 | BIRC6 | 7864 | 0.051 | 0.3230 | No | ||
| 108 | XRCC5 | 7947 | 0.028 | 0.3186 | No | ||
| 109 | CTGF | 8162 | -0.024 | 0.3071 | No | ||
| 110 | RCC1 | 8282 | -0.055 | 0.3008 | No | ||
| 111 | HSPE1 | 8780 | -0.182 | 0.2745 | No | ||
| 112 | SMARCA5 | 8915 | -0.212 | 0.2679 | No | ||
| 113 | EPS8 | 8977 | -0.225 | 0.2652 | No | ||
| 114 | JAG1 | 9243 | -0.288 | 0.2518 | No | ||
| 115 | LYN | 9318 | -0.303 | 0.2487 | No | ||
| 116 | CDC25C | 9340 | -0.307 | 0.2485 | No | ||
| 117 | MTIF2 | 9612 | -0.370 | 0.2350 | No | ||
| 118 | CORO1C | 9738 | -0.399 | 0.2294 | No | ||
| 119 | IL1RL1 | 9750 | -0.401 | 0.2301 | No | ||
| 120 | PDGFB | 10575 | -0.591 | 0.1872 | No | ||
| 121 | CPT1A | 10586 | -0.594 | 0.1885 | No | ||
| 122 | FZD1 | 10875 | -0.659 | 0.1750 | No | ||
| 123 | FOSL1 | 10910 | -0.666 | 0.1752 | No | ||
| 124 | ERCC3 | 11223 | -0.729 | 0.1605 | No | ||
| 125 | SNIP1 | 11247 | -0.735 | 0.1615 | No | ||
| 126 | ADSL | 11405 | -0.769 | 0.1554 | No | ||
| 127 | CHST2 | 11467 | -0.782 | 0.1545 | No | ||
| 128 | THRAP3 | 11703 | -0.827 | 0.1443 | No | ||
| 129 | TDG | 11796 | -0.846 | 0.1420 | No | ||
| 130 | CDK7 | 11914 | -0.871 | 0.1383 | No | ||
| 131 | CRSP6 | 12202 | -0.926 | 0.1256 | No | ||
| 132 | FBXL7 | 12281 | -0.945 | 0.1243 | No | ||
| 133 | MTMR4 | 12380 | -0.967 | 0.1220 | No | ||
| 134 | IPO7 | 12394 | -0.968 | 0.1243 | No | ||
| 135 | CRK | 12483 | -0.986 | 0.1226 | No | ||
| 136 | BLZF1 | 12667 | -1.025 | 0.1158 | No | ||
| 137 | B4GALT5 | 12707 | -1.034 | 0.1169 | No | ||
| 138 | SLC2A1 | 12750 | -1.041 | 0.1178 | No | ||
| 139 | SFRS9 | 12816 | -1.053 | 0.1176 | No | ||
| 140 | CXXC5 | 13524 | -1.213 | 0.0830 | No | ||
| 141 | PSMA5 | 13803 | -1.283 | 0.0719 | No | ||
| 142 | PLAG1 | 14091 | -1.351 | 0.0605 | No | ||
| 143 | RBM27 | 14092 | -1.351 | 0.0647 | No | ||
| 144 | CDH11 | 14188 | -1.375 | 0.0638 | No | ||
| 145 | PANX1 | 14357 | -1.420 | 0.0591 | No | ||
| 146 | NUFIP1 | 14401 | -1.433 | 0.0612 | No | ||
| 147 | BAG2 | 14773 | -1.526 | 0.0458 | No | ||
| 148 | FKBP1A | 14951 | -1.578 | 0.0411 | No | ||
| 149 | CYHR1 | 15146 | -1.638 | 0.0356 | No | ||
| 150 | ARHGEF2 | 15383 | -1.710 | 0.0281 | No | ||
| 151 | HS3ST3B1 | 16236 | -2.038 | -0.0118 | No | ||
| 152 | LHFPL2 | 16542 | -2.181 | -0.0216 | No | ||
| 153 | RBM14 | 16566 | -2.195 | -0.0160 | No | ||
| 154 | UBE2C | 16584 | -2.206 | -0.0101 | No | ||
| 155 | GCNT1 | 16710 | -2.267 | -0.0099 | No | ||
| 156 | BUB1 | 16725 | -2.277 | -0.0036 | No | ||
| 157 | DLG5 | 16947 | -2.413 | -0.0081 | No | ||
| 158 | PAPPA | 17121 | -2.526 | -0.0097 | No | ||
| 159 | BUB3 | 17137 | -2.536 | -0.0027 | No | ||
| 160 | ARFGAP1 | 17163 | -2.557 | 0.0039 | No | ||
| 161 | FADD | 17370 | -2.718 | 0.0011 | No | ||
| 162 | JMJD2B | 17611 | -2.923 | -0.0029 | No | ||
| 163 | GIPC1 | 17639 | -2.963 | 0.0048 | No | ||
| 164 | KIAA0859 | 17823 | -3.152 | 0.0046 | No | ||
| 165 | MID1IP1 | 17920 | -3.283 | 0.0096 | No | ||
| 166 | ZDHHC16 | 18327 | -4.162 | 0.0005 | No | ||
| 167 | WNT5B | 18492 | -4.904 | 0.0067 | No |
