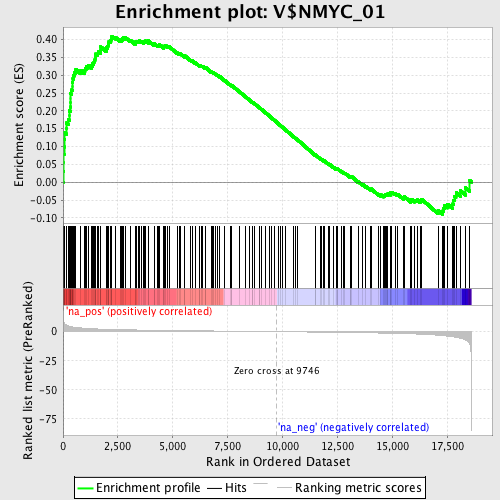
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$NMYC_01 |
| Enrichment Score (ES) | 0.4095072 |
| Normalized Enrichment Score (NES) | 1.8263218 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009564091 |
| FWER p-Value | 0.077 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FBL | 8 | 10.854 | 0.0310 | Yes | ||
| 2 | VARS2 | 25 | 8.561 | 0.0549 | Yes | ||
| 3 | CEBPB | 31 | 8.104 | 0.0781 | Yes | ||
| 4 | HMOX1 | 47 | 7.454 | 0.0989 | Yes | ||
| 5 | TIMM10 | 55 | 7.234 | 0.1194 | Yes | ||
| 6 | ATAD3A | 70 | 6.882 | 0.1386 | Yes | ||
| 7 | ATP5G1 | 152 | 5.608 | 0.1505 | Yes | ||
| 8 | PPAT | 158 | 5.554 | 0.1663 | Yes | ||
| 9 | G6PC3 | 248 | 4.862 | 0.1755 | Yes | ||
| 10 | BCL7C | 287 | 4.642 | 0.1869 | Yes | ||
| 11 | EIF4B | 291 | 4.621 | 0.2001 | Yes | ||
| 12 | NOL5A | 328 | 4.416 | 0.2110 | Yes | ||
| 13 | RCL1 | 336 | 4.377 | 0.2233 | Yes | ||
| 14 | PRDX4 | 340 | 4.362 | 0.2357 | Yes | ||
| 15 | PA2G4 | 341 | 4.356 | 0.2483 | Yes | ||
| 16 | PABPC4 | 378 | 4.228 | 0.2586 | Yes | ||
| 17 | PABPC1 | 419 | 4.086 | 0.2683 | Yes | ||
| 18 | NR1D1 | 421 | 4.076 | 0.2800 | Yes | ||
| 19 | HSPD1 | 446 | 3.986 | 0.2903 | Yes | ||
| 20 | IPO4 | 475 | 3.905 | 0.3001 | Yes | ||
| 21 | SEMA3F | 521 | 3.792 | 0.3086 | Yes | ||
| 22 | CTSF | 579 | 3.645 | 0.3161 | Yes | ||
| 23 | SRP72 | 805 | 3.233 | 0.3132 | Yes | ||
| 24 | AMPD2 | 980 | 2.947 | 0.3123 | Yes | ||
| 25 | HSPE1 | 998 | 2.929 | 0.3199 | Yes | ||
| 26 | MTHFD1 | 1052 | 2.871 | 0.3253 | Yes | ||
| 27 | GPS1 | 1151 | 2.730 | 0.3279 | Yes | ||
| 28 | LMNB1 | 1314 | 2.529 | 0.3265 | Yes | ||
| 29 | UBE1 | 1345 | 2.502 | 0.3321 | Yes | ||
| 30 | GTPBP1 | 1400 | 2.448 | 0.3362 | Yes | ||
| 31 | POLR3E | 1412 | 2.438 | 0.3427 | Yes | ||
| 32 | ELOVL6 | 1455 | 2.393 | 0.3473 | Yes | ||
| 33 | RPL22 | 1476 | 2.371 | 0.3531 | Yes | ||
| 34 | TIAL1 | 1486 | 2.357 | 0.3595 | Yes | ||
| 35 | SFXN2 | 1586 | 2.276 | 0.3607 | Yes | ||
| 36 | HYAL2 | 1592 | 2.273 | 0.3670 | Yes | ||
| 37 | SH3KBP1 | 1696 | 2.201 | 0.3678 | Yes | ||
| 38 | KRTCAP2 | 1711 | 2.189 | 0.3734 | Yes | ||
| 39 | FKBP10 | 1714 | 2.186 | 0.3796 | Yes | ||
| 40 | TPM2 | 1965 | 2.030 | 0.3719 | Yes | ||
| 41 | CTBP2 | 1971 | 2.028 | 0.3775 | Yes | ||
| 42 | CACNA1D | 2016 | 2.004 | 0.3809 | Yes | ||
| 43 | SOX12 | 2048 | 1.987 | 0.3850 | Yes | ||
| 44 | U2AF2 | 2070 | 1.972 | 0.3896 | Yes | ||
| 45 | ZZZ3 | 2088 | 1.960 | 0.3943 | Yes | ||
| 46 | B4GALT2 | 2149 | 1.927 | 0.3967 | Yes | ||
| 47 | ETV1 | 2183 | 1.911 | 0.4004 | Yes | ||
| 48 | GADD45G | 2212 | 1.894 | 0.4044 | Yes | ||
| 49 | LIG3 | 2219 | 1.890 | 0.4095 | Yes | ||
| 50 | SC5DL | 2373 | 1.815 | 0.4065 | No | ||
| 51 | FHOD1 | 2618 | 1.704 | 0.3982 | No | ||
| 52 | TEF | 2673 | 1.680 | 0.4001 | No | ||
| 53 | COPS7A | 2711 | 1.662 | 0.4029 | No | ||
| 54 | TOP1 | 2747 | 1.646 | 0.4058 | No | ||
| 55 | GPD1 | 2848 | 1.606 | 0.4050 | No | ||
| 56 | PES1 | 3064 | 1.522 | 0.3978 | No | ||
| 57 | EIF4G1 | 3294 | 1.441 | 0.3895 | No | ||
| 58 | POGK | 3300 | 1.437 | 0.3934 | No | ||
| 59 | RUNX2 | 3357 | 1.416 | 0.3945 | No | ||
| 60 | RBBP6 | 3422 | 1.390 | 0.3950 | No | ||
| 61 | SLC12A5 | 3459 | 1.379 | 0.3971 | No | ||
| 62 | SOCS5 | 3558 | 1.346 | 0.3956 | No | ||
| 63 | HTF9C | 3683 | 1.304 | 0.3927 | No | ||
| 64 | DLX1 | 3714 | 1.294 | 0.3948 | No | ||
| 65 | BAI2 | 3759 | 1.282 | 0.3961 | No | ||
| 66 | HRSP12 | 3879 | 1.242 | 0.3933 | No | ||
| 67 | HMGN2 | 3884 | 1.241 | 0.3967 | No | ||
| 68 | SLC17A2 | 4144 | 1.167 | 0.3860 | No | ||
| 69 | XRN2 | 4164 | 1.162 | 0.3883 | No | ||
| 70 | GPC3 | 4315 | 1.125 | 0.3834 | No | ||
| 71 | ADPRHL1 | 4366 | 1.110 | 0.3839 | No | ||
| 72 | CDC14A | 4380 | 1.107 | 0.3864 | No | ||
| 73 | ZCCHC7 | 4584 | 1.059 | 0.3785 | No | ||
| 74 | FKBP5 | 4585 | 1.059 | 0.3816 | No | ||
| 75 | RTN4RL2 | 4636 | 1.048 | 0.3819 | No | ||
| 76 | NXPH1 | 4647 | 1.045 | 0.3844 | No | ||
| 77 | NRAS | 4773 | 1.011 | 0.3805 | No | ||
| 78 | ADAM10 | 4832 | 0.997 | 0.3803 | No | ||
| 79 | COL2A1 | 5209 | 0.910 | 0.3625 | No | ||
| 80 | STMN4 | 5315 | 0.881 | 0.3594 | No | ||
| 81 | ARRDC3 | 5353 | 0.872 | 0.3599 | No | ||
| 82 | RPL13A | 5528 | 0.833 | 0.3529 | No | ||
| 83 | GJA1 | 5530 | 0.833 | 0.3552 | No | ||
| 84 | NEUROD2 | 5814 | 0.776 | 0.3421 | No | ||
| 85 | TLE3 | 5880 | 0.763 | 0.3408 | No | ||
| 86 | USP2 | 6024 | 0.731 | 0.3352 | No | ||
| 87 | DNMT3A | 6216 | 0.696 | 0.3268 | No | ||
| 88 | PSEN2 | 6237 | 0.691 | 0.3277 | No | ||
| 89 | PPARGC1B | 6312 | 0.674 | 0.3257 | No | ||
| 90 | MYST3 | 6354 | 0.664 | 0.3254 | No | ||
| 91 | PRKCE | 6499 | 0.633 | 0.3194 | No | ||
| 92 | BARHL1 | 6511 | 0.631 | 0.3206 | No | ||
| 93 | DLC1 | 6753 | 0.581 | 0.3092 | No | ||
| 94 | PLA2G4A | 6792 | 0.571 | 0.3088 | No | ||
| 95 | NKX2-2 | 6855 | 0.561 | 0.3071 | No | ||
| 96 | ADAMTS17 | 6964 | 0.537 | 0.3028 | No | ||
| 97 | NRXN1 | 7049 | 0.521 | 0.2998 | No | ||
| 98 | PCDHA10 | 7146 | 0.502 | 0.2960 | No | ||
| 99 | ATP6V0B | 7374 | 0.457 | 0.2850 | No | ||
| 100 | JPH1 | 7623 | 0.409 | 0.2727 | No | ||
| 101 | BMP2K | 7685 | 0.399 | 0.2706 | No | ||
| 102 | NR5A1 | 7693 | 0.397 | 0.2714 | No | ||
| 103 | LIN28 | 8035 | 0.329 | 0.2538 | No | ||
| 104 | DUSP4 | 8297 | 0.283 | 0.2405 | No | ||
| 105 | WEE1 | 8488 | 0.246 | 0.2309 | No | ||
| 106 | PANK3 | 8627 | 0.216 | 0.2240 | No | ||
| 107 | DNAJB9 | 8648 | 0.214 | 0.2236 | No | ||
| 108 | LAP3 | 8720 | 0.202 | 0.2203 | No | ||
| 109 | MAPKAPK3 | 8935 | 0.166 | 0.2092 | No | ||
| 110 | NPM1 | 8937 | 0.166 | 0.2096 | No | ||
| 111 | GRIN2A | 9051 | 0.144 | 0.2039 | No | ||
| 112 | ALX4 | 9226 | 0.112 | 0.1948 | No | ||
| 113 | HS3ST3A1 | 9428 | 0.072 | 0.1841 | No | ||
| 114 | SLC38A5 | 9503 | 0.055 | 0.1802 | No | ||
| 115 | TIMM9 | 9635 | 0.024 | 0.1732 | No | ||
| 116 | FLT3 | 9819 | -0.016 | 0.1633 | No | ||
| 117 | ABCA1 | 9904 | -0.033 | 0.1589 | No | ||
| 118 | GNA13 | 10003 | -0.059 | 0.1537 | No | ||
| 119 | HPCA | 10127 | -0.085 | 0.1473 | No | ||
| 120 | MAX | 10487 | -0.167 | 0.1283 | No | ||
| 121 | TESK2 | 10584 | -0.183 | 0.1236 | No | ||
| 122 | GRK6 | 10681 | -0.202 | 0.1190 | No | ||
| 123 | ARF6 | 11489 | -0.371 | 0.0763 | No | ||
| 124 | EN2 | 11504 | -0.374 | 0.0766 | No | ||
| 125 | POU3F1 | 11725 | -0.424 | 0.0659 | No | ||
| 126 | RXRB | 11782 | -0.435 | 0.0642 | No | ||
| 127 | LHX9 | 11878 | -0.459 | 0.0603 | No | ||
| 128 | HOXC13 | 11913 | -0.466 | 0.0598 | No | ||
| 129 | B3GALT2 | 12101 | -0.506 | 0.0512 | No | ||
| 130 | CHRM1 | 12158 | -0.520 | 0.0496 | No | ||
| 131 | IPO13 | 12335 | -0.565 | 0.0417 | No | ||
| 132 | SNCAIP | 12452 | -0.593 | 0.0372 | No | ||
| 133 | APOA5 | 12505 | -0.604 | 0.0361 | No | ||
| 134 | SGTB | 12513 | -0.607 | 0.0375 | No | ||
| 135 | SEPT3 | 12680 | -0.647 | 0.0303 | No | ||
| 136 | NUP153 | 12795 | -0.676 | 0.0261 | No | ||
| 137 | KIAA0427 | 12843 | -0.689 | 0.0256 | No | ||
| 138 | RAMP2 | 13084 | -0.756 | 0.0147 | No | ||
| 139 | CHST11 | 13110 | -0.763 | 0.0156 | No | ||
| 140 | FGF11 | 13142 | -0.773 | 0.0161 | No | ||
| 141 | RNF128 | 13442 | -0.851 | 0.0024 | No | ||
| 142 | EIF3S1 | 13663 | -0.919 | -0.0069 | No | ||
| 143 | GADD45B | 13797 | -0.961 | -0.0113 | No | ||
| 144 | SCFD2 | 13999 | -1.030 | -0.0192 | No | ||
| 145 | PFN1 | 14045 | -1.047 | -0.0186 | No | ||
| 146 | ANGPT2 | 14385 | -1.162 | -0.0337 | No | ||
| 147 | NDUFS1 | 14487 | -1.202 | -0.0357 | No | ||
| 148 | MNT | 14597 | -1.236 | -0.0380 | No | ||
| 149 | NEUROD6 | 14647 | -1.254 | -0.0370 | No | ||
| 150 | SEMA7A | 14671 | -1.263 | -0.0346 | No | ||
| 151 | ANXA6 | 14724 | -1.287 | -0.0337 | No | ||
| 152 | SCYL1 | 14786 | -1.309 | -0.0332 | No | ||
| 153 | SLC20A1 | 14804 | -1.317 | -0.0303 | No | ||
| 154 | PLAG1 | 14914 | -1.364 | -0.0323 | No | ||
| 155 | PLAGL1 | 14924 | -1.368 | -0.0288 | No | ||
| 156 | CLCN2 | 14988 | -1.394 | -0.0282 | No | ||
| 157 | HHIP | 15129 | -1.456 | -0.0316 | No | ||
| 158 | ADCY3 | 15261 | -1.521 | -0.0343 | No | ||
| 159 | MCM8 | 15507 | -1.647 | -0.0428 | No | ||
| 160 | ZHX2 | 15549 | -1.666 | -0.0402 | No | ||
| 161 | AKAP10 | 15845 | -1.846 | -0.0508 | No | ||
| 162 | HOXB7 | 15886 | -1.873 | -0.0476 | No | ||
| 163 | MXD4 | 16037 | -1.961 | -0.0500 | No | ||
| 164 | PFKFB3 | 16136 | -2.044 | -0.0494 | No | ||
| 165 | ENPP6 | 16300 | -2.184 | -0.0520 | No | ||
| 166 | HRH3 | 16331 | -2.214 | -0.0472 | No | ||
| 167 | KCNK5 | 17088 | -3.050 | -0.0793 | No | ||
| 168 | RBM3 | 17295 | -3.388 | -0.0807 | No | ||
| 169 | VPS16 | 17345 | -3.485 | -0.0733 | No | ||
| 170 | RAD9A | 17375 | -3.542 | -0.0646 | No | ||
| 171 | AP1GBP1 | 17519 | -3.860 | -0.0612 | No | ||
| 172 | CUGBP1 | 17752 | -4.386 | -0.0610 | No | ||
| 173 | WBP2 | 17796 | -4.515 | -0.0503 | No | ||
| 174 | RPIA | 17834 | -4.622 | -0.0389 | No | ||
| 175 | STAT3 | 17907 | -4.850 | -0.0288 | No | ||
| 176 | PDK2 | 18108 | -5.558 | -0.0235 | No | ||
| 177 | HBP1 | 18344 | -7.095 | -0.0157 | No | ||
| 178 | BCL2 | 18536 | -10.511 | 0.0043 | No |
