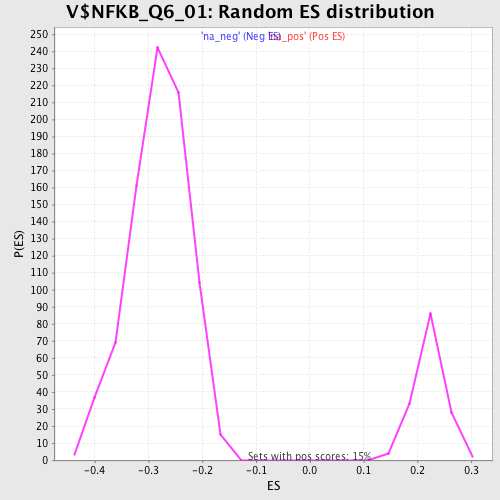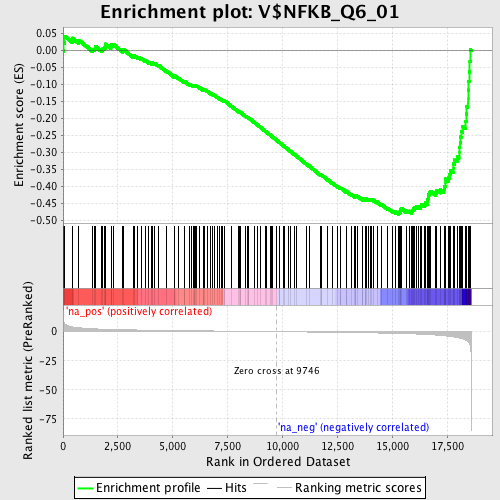
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKB_Q6_01 |
| Enrichment Score (ES) | -0.4817887 |
| Normalized Enrichment Score (NES) | -1.718841 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.085144736 |
| FWER p-Value | 0.769 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF5A | 26 | 8.461 | 0.0246 | No | ||
| 2 | PCBP4 | 77 | 6.779 | 0.0428 | No | ||
| 3 | ARHGAP8 | 412 | 4.098 | 0.0373 | No | ||
| 4 | TCEA2 | 719 | 3.379 | 0.0311 | No | ||
| 5 | GATA4 | 1361 | 2.484 | 0.0040 | No | ||
| 6 | CSF3 | 1452 | 2.398 | 0.0065 | No | ||
| 7 | COL27A1 | 1485 | 2.357 | 0.0120 | No | ||
| 8 | NR3C1 | 1754 | 2.156 | 0.0041 | No | ||
| 9 | NFKBIB | 1814 | 2.114 | 0.0074 | No | ||
| 10 | AMOTL1 | 1885 | 2.075 | 0.0100 | No | ||
| 11 | BMF | 1934 | 2.045 | 0.0137 | No | ||
| 12 | SLC6A12 | 1941 | 2.043 | 0.0197 | No | ||
| 13 | ETV1 | 2183 | 1.911 | 0.0125 | No | ||
| 14 | SEC61A1 | 2190 | 1.906 | 0.0180 | No | ||
| 15 | VCAM1 | 2292 | 1.848 | 0.0182 | No | ||
| 16 | TNFRSF1B | 2726 | 1.655 | -0.0001 | No | ||
| 17 | TOP1 | 2747 | 1.646 | 0.0038 | No | ||
| 18 | TNFSF18 | 3212 | 1.469 | -0.0168 | No | ||
| 19 | CHD4 | 3237 | 1.460 | -0.0136 | No | ||
| 20 | PRKCD | 3412 | 1.394 | -0.0187 | No | ||
| 21 | NTN1 | 3549 | 1.350 | -0.0220 | No | ||
| 22 | BAI2 | 3759 | 1.282 | -0.0293 | No | ||
| 23 | EGF | 3908 | 1.234 | -0.0336 | No | ||
| 24 | DCAMKL1 | 4021 | 1.198 | -0.0359 | No | ||
| 25 | CYLD | 4057 | 1.190 | -0.0342 | No | ||
| 26 | FTHL17 | 4182 | 1.157 | -0.0373 | No | ||
| 27 | ENO3 | 4340 | 1.115 | -0.0424 | No | ||
| 28 | IL12A | 4718 | 1.024 | -0.0597 | No | ||
| 29 | CREB1 | 5058 | 0.945 | -0.0752 | No | ||
| 30 | DDR1 | 5075 | 0.941 | -0.0731 | No | ||
| 31 | AP1S2 | 5265 | 0.892 | -0.0806 | No | ||
| 32 | TRIM47 | 5527 | 0.833 | -0.0922 | No | ||
| 33 | IFNB1 | 5531 | 0.833 | -0.0898 | No | ||
| 34 | UBE2D3 | 5759 | 0.787 | -0.0997 | No | ||
| 35 | SNAP25 | 5828 | 0.775 | -0.1010 | No | ||
| 36 | REV3L | 5922 | 0.754 | -0.1037 | No | ||
| 37 | EDN2 | 5968 | 0.744 | -0.1039 | No | ||
| 38 | BCL6B | 5986 | 0.739 | -0.1025 | No | ||
| 39 | RAP2C | 6022 | 0.732 | -0.1021 | No | ||
| 40 | SMPD3 | 6101 | 0.718 | -0.1042 | No | ||
| 41 | CNTNAP1 | 6234 | 0.693 | -0.1092 | No | ||
| 42 | HSD11B2 | 6401 | 0.653 | -0.1162 | No | ||
| 43 | RASGRP4 | 6402 | 0.653 | -0.1142 | No | ||
| 44 | DAP3 | 6423 | 0.650 | -0.1132 | No | ||
| 45 | PTGES | 6576 | 0.620 | -0.1196 | No | ||
| 46 | UPF2 | 6728 | 0.587 | -0.1260 | No | ||
| 47 | TP53 | 6813 | 0.567 | -0.1288 | No | ||
| 48 | NDRG2 | 6880 | 0.555 | -0.1306 | No | ||
| 49 | RBMS1 | 7038 | 0.523 | -0.1375 | No | ||
| 50 | SHOX2 | 7131 | 0.505 | -0.1410 | No | ||
| 51 | CLCN1 | 7197 | 0.492 | -0.1430 | No | ||
| 52 | PHOX2B | 7280 | 0.477 | -0.1459 | No | ||
| 53 | IL13 | 7352 | 0.461 | -0.1484 | No | ||
| 54 | MOBKL2C | 7369 | 0.458 | -0.1478 | No | ||
| 55 | BMP2K | 7685 | 0.399 | -0.1637 | No | ||
| 56 | IL7 | 7998 | 0.336 | -0.1795 | No | ||
| 57 | RIN2 | 8017 | 0.332 | -0.1795 | No | ||
| 58 | HTR3B | 8082 | 0.320 | -0.1820 | No | ||
| 59 | IL27RA | 8092 | 0.319 | -0.1815 | No | ||
| 60 | RALGDS | 8318 | 0.280 | -0.1928 | No | ||
| 61 | MSX1 | 8381 | 0.266 | -0.1954 | No | ||
| 62 | PTPRJ | 8443 | 0.256 | -0.1979 | No | ||
| 63 | POU2F3 | 8465 | 0.253 | -0.1982 | No | ||
| 64 | TNFSF15 | 8700 | 0.205 | -0.2103 | No | ||
| 65 | BCL3 | 8868 | 0.178 | -0.2188 | No | ||
| 66 | MADCAM1 | 8991 | 0.156 | -0.2249 | No | ||
| 67 | FGF17 | 9220 | 0.113 | -0.2369 | No | ||
| 68 | KRTAP13-1 | 9272 | 0.103 | -0.2394 | No | ||
| 69 | ZBTB11 | 9430 | 0.071 | -0.2477 | No | ||
| 70 | HSD3B7 | 9512 | 0.053 | -0.2519 | No | ||
| 71 | SIRT2 | 9552 | 0.044 | -0.2539 | No | ||
| 72 | IER5 | 9740 | 0.002 | -0.2640 | No | ||
| 73 | RND1 | 9859 | -0.024 | -0.2703 | No | ||
| 74 | IL27 | 9873 | -0.027 | -0.2709 | No | ||
| 75 | WNT10A | 10027 | -0.064 | -0.2790 | No | ||
| 76 | SIN3A | 10072 | -0.073 | -0.2812 | No | ||
| 77 | PTHLH | 10250 | -0.112 | -0.2904 | No | ||
| 78 | MSC | 10353 | -0.137 | -0.2955 | No | ||
| 79 | KLK9 | 10555 | -0.178 | -0.3059 | No | ||
| 80 | IL1RN | 10641 | -0.195 | -0.3099 | No | ||
| 81 | C1QL1 | 11106 | -0.292 | -0.3341 | No | ||
| 82 | PAPPA | 11212 | -0.313 | -0.3389 | No | ||
| 83 | RFX5 | 11711 | -0.420 | -0.3646 | No | ||
| 84 | EHF | 11755 | -0.429 | -0.3656 | No | ||
| 85 | MAG | 11791 | -0.438 | -0.3661 | No | ||
| 86 | EBI3 | 12066 | -0.497 | -0.3794 | No | ||
| 87 | SEMA3B | 12298 | -0.556 | -0.3902 | No | ||
| 88 | ERN1 | 12523 | -0.609 | -0.4005 | No | ||
| 89 | BATF | 12636 | -0.636 | -0.4046 | No | ||
| 90 | GABRB1 | 12663 | -0.643 | -0.4041 | No | ||
| 91 | JAK3 | 12893 | -0.700 | -0.4143 | No | ||
| 92 | BDNF | 12902 | -0.703 | -0.4126 | No | ||
| 93 | GREM1 | 13152 | -0.775 | -0.4237 | No | ||
| 94 | DUSP22 | 13301 | -0.811 | -0.4292 | No | ||
| 95 | NEK8 | 13305 | -0.813 | -0.4269 | No | ||
| 96 | EDG5 | 13319 | -0.815 | -0.4251 | No | ||
| 97 | ALG6 | 13420 | -0.844 | -0.4279 | No | ||
| 98 | MITF | 13644 | -0.915 | -0.4372 | No | ||
| 99 | GNG4 | 13666 | -0.920 | -0.4355 | No | ||
| 100 | SOX5 | 13777 | -0.957 | -0.4385 | No | ||
| 101 | GADD45B | 13797 | -0.961 | -0.4366 | No | ||
| 102 | IL23A | 13841 | -0.977 | -0.4359 | No | ||
| 103 | SLAMF8 | 13936 | -1.005 | -0.4379 | No | ||
| 104 | NR2F2 | 14000 | -1.030 | -0.4381 | No | ||
| 105 | PFN1 | 14045 | -1.047 | -0.4373 | No | ||
| 106 | ASCL3 | 14140 | -1.077 | -0.4391 | No | ||
| 107 | TCF1 | 14307 | -1.137 | -0.4446 | No | ||
| 108 | GRK5 | 14502 | -1.205 | -0.4514 | No | ||
| 109 | TSLP | 14787 | -1.309 | -0.4627 | No | ||
| 110 | FGF1 | 15032 | -1.413 | -0.4716 | No | ||
| 111 | UBE2H | 15161 | -1.470 | -0.4740 | No | ||
| 112 | GNB1 | 15305 | -1.543 | -0.4770 | Yes | ||
| 113 | BIRC3 | 15345 | -1.560 | -0.4744 | Yes | ||
| 114 | ARHGEF2 | 15378 | -1.586 | -0.4712 | Yes | ||
| 115 | UBE2I | 15387 | -1.589 | -0.4668 | Yes | ||
| 116 | FUT7 | 15409 | -1.598 | -0.4630 | Yes | ||
| 117 | CENTB5 | 15630 | -1.712 | -0.4696 | Yes | ||
| 118 | STAT6 | 15767 | -1.794 | -0.4715 | Yes | ||
| 119 | ICAM1 | 15892 | -1.876 | -0.4724 | Yes | ||
| 120 | PCSK2 | 15930 | -1.899 | -0.4686 | Yes | ||
| 121 | SP6 | 15979 | -1.922 | -0.4653 | Yes | ||
| 122 | NLK | 16010 | -1.942 | -0.4609 | Yes | ||
| 123 | UACA | 16097 | -2.000 | -0.4594 | Yes | ||
| 124 | GPHN | 16180 | -2.081 | -0.4575 | Yes | ||
| 125 | IL2RA | 16308 | -2.190 | -0.4576 | Yes | ||
| 126 | TRIB2 | 16336 | -2.218 | -0.4523 | Yes | ||
| 127 | BLR1 | 16474 | -2.336 | -0.4525 | Yes | ||
| 128 | ILK | 16520 | -2.381 | -0.4476 | Yes | ||
| 129 | RELB | 16614 | -2.472 | -0.4451 | Yes | ||
| 130 | GRIN2D | 16618 | -2.475 | -0.4376 | Yes | ||
| 131 | HIVEP1 | 16636 | -2.491 | -0.4309 | Yes | ||
| 132 | ASH1L | 16643 | -2.501 | -0.4235 | Yes | ||
| 133 | NFKBIA | 16685 | -2.540 | -0.4179 | Yes | ||
| 134 | TBX20 | 16739 | -2.604 | -0.4128 | Yes | ||
| 135 | NFKB2 | 16979 | -2.902 | -0.4168 | Yes | ||
| 136 | MRPS6 | 17022 | -2.955 | -0.4100 | Yes | ||
| 137 | MAP3K11 | 17183 | -3.201 | -0.4088 | Yes | ||
| 138 | ATP1B1 | 17361 | -3.522 | -0.4076 | Yes | ||
| 139 | ELMO1 | 17391 | -3.567 | -0.3982 | Yes | ||
| 140 | SOCS2 | 17411 | -3.609 | -0.3881 | Yes | ||
| 141 | MMP9 | 17416 | -3.617 | -0.3772 | Yes | ||
| 142 | IL6ST | 17562 | -3.942 | -0.3729 | Yes | ||
| 143 | RHOG | 17604 | -4.011 | -0.3628 | Yes | ||
| 144 | IL4I1 | 17640 | -4.117 | -0.3520 | Yes | ||
| 145 | NFAT5 | 17779 | -4.490 | -0.3457 | Yes | ||
| 146 | XPO6 | 17786 | -4.501 | -0.3322 | Yes | ||
| 147 | LTA | 17854 | -4.683 | -0.3214 | Yes | ||
| 148 | TNFRSF9 | 17957 | -4.990 | -0.3116 | Yes | ||
| 149 | ZNRF1 | 18044 | -5.291 | -0.2999 | Yes | ||
| 150 | PPP1R13B | 18047 | -5.296 | -0.2838 | Yes | ||
| 151 | SMOC1 | 18089 | -5.468 | -0.2692 | Yes | ||
| 152 | ZIC4 | 18117 | -5.585 | -0.2534 | Yes | ||
| 153 | RASSF2 | 18154 | -5.765 | -0.2377 | Yes | ||
| 154 | USP52 | 18212 | -6.083 | -0.2220 | Yes | ||
| 155 | UBD | 18354 | -7.211 | -0.2075 | Yes | ||
| 156 | TNIP1 | 18372 | -7.332 | -0.1859 | Yes | ||
| 157 | CD69 | 18401 | -7.609 | -0.1640 | Yes | ||
| 158 | TRAF4 | 18453 | -8.305 | -0.1412 | Yes | ||
| 159 | MAP4K2 | 18455 | -8.325 | -0.1156 | Yes | ||
| 160 | FBXL12 | 18486 | -8.863 | -0.0900 | Yes | ||
| 161 | LTB | 18510 | -9.387 | -0.0624 | Yes | ||
| 162 | DUSP6 | 18542 | -10.788 | -0.0308 | Yes | ||
| 163 | CCL5 | 18555 | -11.313 | 0.0033 | Yes |