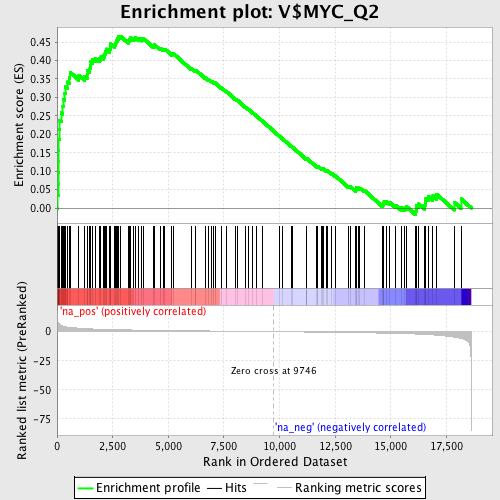
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.46622148 |
| Normalized Enrichment Score (NES) | 1.9635048 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.010519054 |
| FWER p-Value | 0.022 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TAGLN2 | 30 | 8.107 | 0.0344 | Yes | ||
| 2 | TIMM10 | 55 | 7.234 | 0.0653 | Yes | ||
| 3 | SHMT2 | 58 | 7.173 | 0.0971 | Yes | ||
| 4 | ATAD3A | 70 | 6.882 | 0.1271 | Yes | ||
| 5 | PFDN2 | 74 | 6.806 | 0.1572 | Yes | ||
| 6 | HNRPA1 | 80 | 6.737 | 0.1869 | Yes | ||
| 7 | ADK | 101 | 6.219 | 0.2135 | Yes | ||
| 8 | NUDC | 120 | 5.874 | 0.2387 | Yes | ||
| 9 | LDHA | 184 | 5.314 | 0.2589 | Yes | ||
| 10 | EBNA1BP2 | 261 | 4.764 | 0.2760 | Yes | ||
| 11 | EIF4B | 291 | 4.621 | 0.2950 | Yes | ||
| 12 | PA2G4 | 341 | 4.356 | 0.3117 | Yes | ||
| 13 | PABPC4 | 378 | 4.228 | 0.3286 | Yes | ||
| 14 | CFL1 | 458 | 3.955 | 0.3419 | Yes | ||
| 15 | AK2 | 542 | 3.762 | 0.3541 | Yes | ||
| 16 | CTSF | 579 | 3.645 | 0.3684 | Yes | ||
| 17 | AMPD2 | 980 | 2.947 | 0.3599 | Yes | ||
| 18 | IVNS1ABP | 1243 | 2.607 | 0.3573 | Yes | ||
| 19 | FKBP11 | 1351 | 2.494 | 0.3626 | Yes | ||
| 20 | EIF3S10 | 1365 | 2.478 | 0.3730 | Yes | ||
| 21 | RPL22 | 1476 | 2.371 | 0.3776 | Yes | ||
| 22 | TIAL1 | 1486 | 2.357 | 0.3876 | Yes | ||
| 23 | RAB3IL1 | 1514 | 2.334 | 0.3965 | Yes | ||
| 24 | SFXN2 | 1586 | 2.276 | 0.4028 | Yes | ||
| 25 | KRTCAP2 | 1711 | 2.189 | 0.4058 | Yes | ||
| 26 | PPRC1 | 1892 | 2.070 | 0.4053 | Yes | ||
| 27 | CTBP2 | 1971 | 2.028 | 0.4101 | Yes | ||
| 28 | ZZZ3 | 2088 | 1.960 | 0.4125 | Yes | ||
| 29 | B4GALT2 | 2149 | 1.927 | 0.4179 | Yes | ||
| 30 | GTF2H1 | 2175 | 1.914 | 0.4250 | Yes | ||
| 31 | AP3M1 | 2215 | 1.893 | 0.4313 | Yes | ||
| 32 | SC5DL | 2373 | 1.815 | 0.4309 | Yes | ||
| 33 | TFB2M | 2387 | 1.807 | 0.4383 | Yes | ||
| 34 | ATP5F1 | 2408 | 1.794 | 0.4452 | Yes | ||
| 35 | UBXD3 | 2601 | 1.714 | 0.4424 | Yes | ||
| 36 | FABP3 | 2644 | 1.693 | 0.4477 | Yes | ||
| 37 | STMN1 | 2649 | 1.690 | 0.4550 | Yes | ||
| 38 | COPS7A | 2711 | 1.662 | 0.4591 | Yes | ||
| 39 | RHEBL1 | 2751 | 1.643 | 0.4643 | Yes | ||
| 40 | GPD1 | 2848 | 1.606 | 0.4662 | Yes | ||
| 41 | LTBR | 3198 | 1.474 | 0.4539 | No | ||
| 42 | CHD4 | 3237 | 1.460 | 0.4584 | No | ||
| 43 | POGK | 3300 | 1.437 | 0.4614 | No | ||
| 44 | COMMD3 | 3444 | 1.382 | 0.4598 | No | ||
| 45 | ALDH3B1 | 3522 | 1.358 | 0.4617 | No | ||
| 46 | PITX3 | 3657 | 1.311 | 0.4603 | No | ||
| 47 | SYT6 | 3793 | 1.270 | 0.4586 | No | ||
| 48 | HMGN2 | 3884 | 1.241 | 0.4593 | No | ||
| 49 | AGMAT | 4337 | 1.116 | 0.4398 | No | ||
| 50 | CDC14A | 4380 | 1.107 | 0.4425 | No | ||
| 51 | RTN4RL2 | 4636 | 1.048 | 0.4333 | No | ||
| 52 | NRAS | 4773 | 1.011 | 0.4305 | No | ||
| 53 | CNNM1 | 4845 | 0.994 | 0.4311 | No | ||
| 54 | TAF6L | 5153 | 0.923 | 0.4186 | No | ||
| 55 | COL2A1 | 5209 | 0.910 | 0.4197 | No | ||
| 56 | USP2 | 6024 | 0.731 | 0.3789 | No | ||
| 57 | HNRPF | 6202 | 0.698 | 0.3724 | No | ||
| 58 | PSEN2 | 6237 | 0.691 | 0.3737 | No | ||
| 59 | H3F3A | 6649 | 0.605 | 0.3542 | No | ||
| 60 | PLA2G4A | 6792 | 0.571 | 0.3490 | No | ||
| 61 | CGN | 6929 | 0.544 | 0.3441 | No | ||
| 62 | SLC16A1 | 7026 | 0.525 | 0.3412 | No | ||
| 63 | FOXD3 | 7106 | 0.510 | 0.3392 | No | ||
| 64 | ATP6V0B | 7374 | 0.457 | 0.3268 | No | ||
| 65 | HOXC12 | 7600 | 0.412 | 0.3165 | No | ||
| 66 | LIN28 | 8035 | 0.329 | 0.2945 | No | ||
| 67 | SIRT1 | 8112 | 0.315 | 0.2918 | No | ||
| 68 | WEE1 | 8488 | 0.246 | 0.2726 | No | ||
| 69 | SLC6A15 | 8598 | 0.223 | 0.2677 | No | ||
| 70 | FADS3 | 8794 | 0.189 | 0.2580 | No | ||
| 71 | SLC1A7 | 8982 | 0.157 | 0.2486 | No | ||
| 72 | ALX4 | 9226 | 0.112 | 0.2360 | No | ||
| 73 | BHLHB3 | 9995 | -0.058 | 0.1947 | No | ||
| 74 | HPCA | 10127 | -0.085 | 0.1880 | No | ||
| 75 | OPRD1 | 10533 | -0.175 | 0.1669 | No | ||
| 76 | TESK2 | 10584 | -0.183 | 0.1650 | No | ||
| 77 | RRAGC | 11205 | -0.311 | 0.1329 | No | ||
| 78 | PAX6 | 11207 | -0.311 | 0.1342 | No | ||
| 79 | TGFB2 | 11211 | -0.313 | 0.1354 | No | ||
| 80 | IPO7 | 11662 | -0.410 | 0.1129 | No | ||
| 81 | RFX5 | 11711 | -0.420 | 0.1122 | No | ||
| 82 | POU3F1 | 11725 | -0.424 | 0.1134 | No | ||
| 83 | LHX9 | 11878 | -0.459 | 0.1072 | No | ||
| 84 | HOXC13 | 11913 | -0.466 | 0.1075 | No | ||
| 85 | USP15 | 11955 | -0.475 | 0.1073 | No | ||
| 86 | B3GALT2 | 12101 | -0.506 | 0.1018 | No | ||
| 87 | CHRM1 | 12158 | -0.520 | 0.1010 | No | ||
| 88 | IPO13 | 12335 | -0.565 | 0.0940 | No | ||
| 89 | APOA5 | 12505 | -0.604 | 0.0876 | No | ||
| 90 | NR0B2 | 13116 | -0.765 | 0.0580 | No | ||
| 91 | FOSL1 | 13166 | -0.779 | 0.0588 | No | ||
| 92 | TDRD1 | 13428 | -0.846 | 0.0485 | No | ||
| 93 | CSDA | 13436 | -0.848 | 0.0519 | No | ||
| 94 | B3GALT6 | 13443 | -0.851 | 0.0553 | No | ||
| 95 | HPCAL4 | 13528 | -0.879 | 0.0547 | No | ||
| 96 | RUSC1 | 13610 | -0.906 | 0.0544 | No | ||
| 97 | LMX1A | 13804 | -0.964 | 0.0482 | No | ||
| 98 | NRIP3 | 14637 | -1.250 | 0.0088 | No | ||
| 99 | TRIM46 | 14653 | -1.256 | 0.0136 | No | ||
| 100 | NKX2-3 | 14692 | -1.273 | 0.0172 | No | ||
| 101 | SCYL1 | 14786 | -1.309 | 0.0180 | No | ||
| 102 | ESRRA | 14952 | -1.382 | 0.0152 | No | ||
| 103 | LRP8 | 15221 | -1.495 | 0.0074 | No | ||
| 104 | FCHSD2 | 15461 | -1.625 | 0.0017 | No | ||
| 105 | CENTB5 | 15630 | -1.712 | 0.0002 | No | ||
| 106 | CBX5 | 15705 | -1.755 | 0.0040 | No | ||
| 107 | BCL9L | 16103 | -2.004 | -0.0085 | No | ||
| 108 | PFKFB3 | 16136 | -2.044 | -0.0012 | No | ||
| 109 | ARL3 | 16165 | -2.072 | 0.0066 | No | ||
| 110 | IL15RA | 16258 | -2.145 | 0.0111 | No | ||
| 111 | ILK | 16520 | -2.381 | 0.0076 | No | ||
| 112 | ATF7IP | 16541 | -2.408 | 0.0172 | No | ||
| 113 | NIT1 | 16577 | -2.439 | 0.0262 | No | ||
| 114 | PTPRF | 16694 | -2.553 | 0.0313 | No | ||
| 115 | FXYD2 | 16892 | -2.803 | 0.0331 | No | ||
| 116 | RORC | 17063 | -3.008 | 0.0373 | No | ||
| 117 | FXYD6 | 17853 | -4.680 | 0.0154 | No | ||
| 118 | TMEM24 | 18156 | -5.788 | 0.0249 | No |
