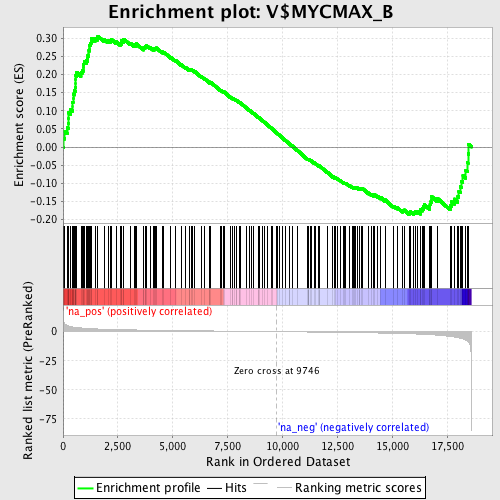
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYCMAX_B |
| Enrichment Score (ES) | 0.30561045 |
| Normalized Enrichment Score (NES) | 1.3487661 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.20242585 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF5A | 26 | 8.461 | 0.0246 | Yes | ||
| 2 | PCBP4 | 77 | 6.779 | 0.0427 | Yes | ||
| 3 | EIF3S9 | 187 | 5.303 | 0.0530 | Yes | ||
| 4 | SNRP70 | 237 | 4.903 | 0.0654 | Yes | ||
| 5 | G6PC3 | 248 | 4.862 | 0.0798 | Yes | ||
| 6 | YTHDF2 | 255 | 4.819 | 0.0943 | Yes | ||
| 7 | RCL1 | 336 | 4.377 | 0.1034 | Yes | ||
| 8 | PABPC1 | 419 | 4.086 | 0.1115 | Yes | ||
| 9 | NR1D1 | 421 | 4.076 | 0.1239 | Yes | ||
| 10 | RPS19 | 455 | 3.961 | 0.1343 | Yes | ||
| 11 | EXOSC5 | 460 | 3.949 | 0.1462 | Yes | ||
| 12 | CEBPA | 514 | 3.808 | 0.1550 | Yes | ||
| 13 | EBAG9 | 554 | 3.730 | 0.1643 | Yes | ||
| 14 | SPG21 | 569 | 3.677 | 0.1749 | Yes | ||
| 15 | NUTF2 | 572 | 3.669 | 0.1860 | Yes | ||
| 16 | MYOHD1 | 577 | 3.646 | 0.1970 | Yes | ||
| 17 | GTF2A1 | 621 | 3.547 | 0.2056 | Yes | ||
| 18 | CRK | 816 | 3.207 | 0.2049 | Yes | ||
| 19 | RNF125 | 865 | 3.132 | 0.2119 | Yes | ||
| 20 | DNAJC11 | 916 | 3.048 | 0.2185 | Yes | ||
| 21 | SDHB | 921 | 3.041 | 0.2276 | Yes | ||
| 22 | RAI14 | 952 | 2.986 | 0.2352 | Yes | ||
| 23 | ETF1 | 1065 | 2.849 | 0.2379 | Yes | ||
| 24 | RANBP1 | 1098 | 2.805 | 0.2447 | Yes | ||
| 25 | ACY1 | 1128 | 2.764 | 0.2517 | Yes | ||
| 26 | PHF7 | 1173 | 2.703 | 0.2576 | Yes | ||
| 27 | NCL | 1174 | 2.702 | 0.2659 | Yes | ||
| 28 | SDK1 | 1202 | 2.659 | 0.2726 | Yes | ||
| 29 | SYT7 | 1205 | 2.653 | 0.2806 | Yes | ||
| 30 | IVNS1ABP | 1243 | 2.607 | 0.2866 | Yes | ||
| 31 | VAPA | 1277 | 2.561 | 0.2927 | Yes | ||
| 32 | RBBP7 | 1300 | 2.547 | 0.2993 | Yes | ||
| 33 | ELOVL6 | 1455 | 2.393 | 0.2983 | Yes | ||
| 34 | CXCL14 | 1550 | 2.302 | 0.3002 | Yes | ||
| 35 | PER1 | 1581 | 2.278 | 0.3056 | Yes | ||
| 36 | RPS6KA3 | 1863 | 2.086 | 0.2968 | No | ||
| 37 | SOX12 | 2048 | 1.987 | 0.2929 | No | ||
| 38 | HMGA1 | 2154 | 1.926 | 0.2931 | No | ||
| 39 | SEC61A1 | 2190 | 1.906 | 0.2971 | No | ||
| 40 | EEF1B2 | 2420 | 1.789 | 0.2901 | No | ||
| 41 | UBXD3 | 2601 | 1.714 | 0.2856 | No | ||
| 42 | CCT5 | 2648 | 1.691 | 0.2883 | No | ||
| 43 | STMN1 | 2649 | 1.690 | 0.2935 | No | ||
| 44 | PTK7 | 2748 | 1.644 | 0.2933 | No | ||
| 45 | TNPO2 | 2762 | 1.639 | 0.2976 | No | ||
| 46 | PES1 | 3064 | 1.522 | 0.2859 | No | ||
| 47 | JUB | 3233 | 1.464 | 0.2813 | No | ||
| 48 | HNRPDL | 3313 | 1.432 | 0.2814 | No | ||
| 49 | EGR3 | 3322 | 1.429 | 0.2854 | No | ||
| 50 | PITX3 | 3657 | 1.311 | 0.2713 | No | ||
| 51 | HTF9C | 3683 | 1.304 | 0.2739 | No | ||
| 52 | SIX1 | 3734 | 1.287 | 0.2752 | No | ||
| 53 | CCND1 | 3753 | 1.284 | 0.2781 | No | ||
| 54 | KPNB1 | 3811 | 1.264 | 0.2789 | No | ||
| 55 | HSF4 | 3966 | 1.216 | 0.2743 | No | ||
| 56 | BRUNOL4 | 4114 | 1.177 | 0.2699 | No | ||
| 57 | XRN2 | 4164 | 1.162 | 0.2709 | No | ||
| 58 | ASCL2 | 4198 | 1.153 | 0.2726 | No | ||
| 59 | NFATC4 | 4253 | 1.142 | 0.2732 | No | ||
| 60 | LRFN5 | 4516 | 1.073 | 0.2623 | No | ||
| 61 | PCTK1 | 4587 | 1.058 | 0.2617 | No | ||
| 62 | BAP1 | 4904 | 0.980 | 0.2476 | No | ||
| 63 | HNRPR | 5108 | 0.932 | 0.2394 | No | ||
| 64 | HOXA9 | 5376 | 0.866 | 0.2276 | No | ||
| 65 | TBX2 | 5578 | 0.826 | 0.2193 | No | ||
| 66 | UBE2D3 | 5759 | 0.787 | 0.2119 | No | ||
| 67 | ASCL1 | 5781 | 0.783 | 0.2132 | No | ||
| 68 | SNAP25 | 5828 | 0.775 | 0.2131 | No | ||
| 69 | ANAPC10 | 5891 | 0.760 | 0.2120 | No | ||
| 70 | NR2E1 | 5995 | 0.738 | 0.2087 | No | ||
| 71 | PPARGC1B | 6312 | 0.674 | 0.1936 | No | ||
| 72 | STAG2 | 6458 | 0.642 | 0.1878 | No | ||
| 73 | PICALM | 6681 | 0.598 | 0.1775 | No | ||
| 74 | TAF11 | 6682 | 0.598 | 0.1794 | No | ||
| 75 | EPS8 | 6738 | 0.585 | 0.1782 | No | ||
| 76 | HOXC5 | 7170 | 0.497 | 0.1563 | No | ||
| 77 | PTMA | 7233 | 0.486 | 0.1545 | No | ||
| 78 | PLXNC1 | 7307 | 0.470 | 0.1520 | No | ||
| 79 | NELF | 7337 | 0.464 | 0.1518 | No | ||
| 80 | CPNE5 | 7618 | 0.410 | 0.1379 | No | ||
| 81 | DLL4 | 7723 | 0.390 | 0.1334 | No | ||
| 82 | TCOF1 | 7729 | 0.389 | 0.1344 | No | ||
| 83 | SLC25A5 | 7802 | 0.376 | 0.1316 | No | ||
| 84 | HNRPD | 7882 | 0.358 | 0.1284 | No | ||
| 85 | APLN | 7885 | 0.357 | 0.1294 | No | ||
| 86 | ZDHHC5 | 8027 | 0.331 | 0.1228 | No | ||
| 87 | SERPINF1 | 8042 | 0.328 | 0.1230 | No | ||
| 88 | BZW2 | 8103 | 0.317 | 0.1208 | No | ||
| 89 | ZFP1 | 8358 | 0.270 | 0.1078 | No | ||
| 90 | WEE1 | 8488 | 0.246 | 0.1016 | No | ||
| 91 | NSF | 8577 | 0.227 | 0.0975 | No | ||
| 92 | OSBPL10 | 8680 | 0.208 | 0.0926 | No | ||
| 93 | GRIN2B | 8897 | 0.172 | 0.0814 | No | ||
| 94 | WNT1 | 8919 | 0.169 | 0.0808 | No | ||
| 95 | NPM1 | 8937 | 0.166 | 0.0804 | No | ||
| 96 | HOXC4 | 9075 | 0.139 | 0.0734 | No | ||
| 97 | CRABP2 | 9173 | 0.121 | 0.0685 | No | ||
| 98 | MMP11 | 9310 | 0.094 | 0.0614 | No | ||
| 99 | SYT12 | 9478 | 0.060 | 0.0525 | No | ||
| 100 | CALM2 | 9554 | 0.043 | 0.0486 | No | ||
| 101 | VAX1 | 9724 | 0.004 | 0.0394 | No | ||
| 102 | CATSPER2 | 9770 | -0.006 | 0.0370 | No | ||
| 103 | TRIB1 | 9842 | -0.020 | 0.0332 | No | ||
| 104 | BHLHB3 | 9995 | -0.058 | 0.0252 | No | ||
| 105 | HPCA | 10127 | -0.085 | 0.0183 | No | ||
| 106 | NEUROD1 | 10311 | -0.126 | 0.0088 | No | ||
| 107 | FOXA2 | 10338 | -0.135 | 0.0078 | No | ||
| 108 | SPOP | 10432 | -0.154 | 0.0032 | No | ||
| 109 | ALOXE3 | 10687 | -0.204 | -0.0099 | No | ||
| 110 | SMAD7 | 11130 | -0.297 | -0.0330 | No | ||
| 111 | SCML2 | 11166 | -0.305 | -0.0340 | No | ||
| 112 | RKHD3 | 11185 | -0.308 | -0.0340 | No | ||
| 113 | FGF14 | 11201 | -0.310 | -0.0338 | No | ||
| 114 | ABCE1 | 11265 | -0.325 | -0.0363 | No | ||
| 115 | SNX5 | 11340 | -0.339 | -0.0392 | No | ||
| 116 | KIT | 11438 | -0.360 | -0.0434 | No | ||
| 117 | ARF6 | 11489 | -0.371 | -0.0450 | No | ||
| 118 | PIGW | 11620 | -0.400 | -0.0508 | No | ||
| 119 | IPO7 | 11662 | -0.410 | -0.0518 | No | ||
| 120 | UBE2S | 11704 | -0.419 | -0.0527 | No | ||
| 121 | HOXB9 | 12053 | -0.495 | -0.0700 | No | ||
| 122 | TGIF2 | 12299 | -0.556 | -0.0816 | No | ||
| 123 | NEUROG3 | 12388 | -0.575 | -0.0846 | No | ||
| 124 | PAIP2 | 12408 | -0.581 | -0.0839 | No | ||
| 125 | PTOV1 | 12519 | -0.608 | -0.0880 | No | ||
| 126 | ELAVL3 | 12621 | -0.632 | -0.0915 | No | ||
| 127 | RNF146 | 12775 | -0.671 | -0.0978 | No | ||
| 128 | KIAA0427 | 12843 | -0.689 | -0.0993 | No | ||
| 129 | HN1 | 12890 | -0.700 | -0.0996 | No | ||
| 130 | BCKDHA | 13063 | -0.750 | -0.1066 | No | ||
| 131 | NKX2-8 | 13175 | -0.780 | -0.1103 | No | ||
| 132 | EPHA2 | 13246 | -0.797 | -0.1116 | No | ||
| 133 | SALL1 | 13282 | -0.805 | -0.1110 | No | ||
| 134 | SLC1A1 | 13323 | -0.816 | -0.1107 | No | ||
| 135 | IRX2 | 13405 | -0.841 | -0.1125 | No | ||
| 136 | PTF1A | 13492 | -0.865 | -0.1145 | No | ||
| 137 | HPCAL4 | 13528 | -0.879 | -0.1137 | No | ||
| 138 | SOCS3 | 13595 | -0.902 | -0.1145 | No | ||
| 139 | DAZAP1 | 13639 | -0.914 | -0.1141 | No | ||
| 140 | DLGAP4 | 13924 | -1.001 | -0.1264 | No | ||
| 141 | PFN1 | 14045 | -1.047 | -0.1297 | No | ||
| 142 | ISL1 | 14150 | -1.079 | -0.1320 | No | ||
| 143 | PRKAG2 | 14172 | -1.091 | -0.1298 | No | ||
| 144 | ILF3 | 14335 | -1.146 | -0.1351 | No | ||
| 145 | NDUFS1 | 14487 | -1.202 | -0.1396 | No | ||
| 146 | SEMA7A | 14671 | -1.263 | -0.1456 | No | ||
| 147 | SOX4 | 15077 | -1.432 | -0.1632 | No | ||
| 148 | LRP8 | 15221 | -1.495 | -0.1664 | No | ||
| 149 | FCHSD2 | 15461 | -1.625 | -0.1743 | No | ||
| 150 | ALDH1A2 | 15539 | -1.660 | -0.1734 | No | ||
| 151 | PDGFB | 15770 | -1.798 | -0.1804 | No | ||
| 152 | PLCD1 | 15818 | -1.827 | -0.1773 | No | ||
| 153 | ZDHHC14 | 15974 | -1.917 | -0.1798 | No | ||
| 154 | PAX2 | 16038 | -1.962 | -0.1772 | No | ||
| 155 | ARL3 | 16165 | -2.072 | -0.1777 | No | ||
| 156 | XYLT2 | 16297 | -2.182 | -0.1781 | No | ||
| 157 | ENPP6 | 16300 | -2.184 | -0.1715 | No | ||
| 158 | TRIM8 | 16391 | -2.264 | -0.1694 | No | ||
| 159 | SPAG9 | 16430 | -2.297 | -0.1644 | No | ||
| 160 | GFI1 | 16450 | -2.314 | -0.1584 | No | ||
| 161 | H1FX | 16708 | -2.570 | -0.1644 | No | ||
| 162 | CALM3 | 16720 | -2.583 | -0.1571 | No | ||
| 163 | DAZL | 16763 | -2.637 | -0.1513 | No | ||
| 164 | PIP5K2B | 16781 | -2.656 | -0.1440 | No | ||
| 165 | DBP | 16786 | -2.664 | -0.1361 | No | ||
| 166 | RORC | 17063 | -3.008 | -0.1418 | No | ||
| 167 | INSM1 | 17646 | -4.125 | -0.1607 | No | ||
| 168 | HS3ST3B1 | 17711 | -4.294 | -0.1510 | No | ||
| 169 | ANKMY2 | 17829 | -4.610 | -0.1432 | No | ||
| 170 | KLF13 | 17992 | -5.096 | -0.1363 | No | ||
| 171 | BCL11B | 18037 | -5.272 | -0.1225 | No | ||
| 172 | ZFP36L1 | 18112 | -5.581 | -0.1094 | No | ||
| 173 | SRF | 18153 | -5.754 | -0.0939 | No | ||
| 174 | SFRS14 | 18223 | -6.139 | -0.0788 | No | ||
| 175 | PPP1R9B | 18353 | -7.203 | -0.0637 | No | ||
| 176 | CRMP1 | 18440 | -8.154 | -0.0433 | No | ||
| 177 | KCNH2 | 18459 | -8.368 | -0.0186 | No | ||
| 178 | TRPC4AP | 18483 | -8.827 | 0.0072 | No |
