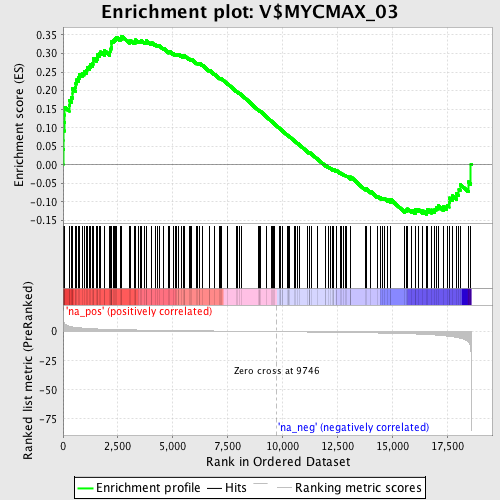
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MYCMAX_03 |
| Enrichment Score (ES) | 0.34691343 |
| Normalized Enrichment Score (NES) | 1.5488393 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10266453 |
| FWER p-Value | 0.74 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BMP7 | 3 | 13.344 | 0.0418 | Yes | ||
| 2 | BAX | 28 | 8.257 | 0.0664 | Yes | ||
| 3 | CEBPB | 31 | 8.104 | 0.0918 | Yes | ||
| 4 | HMOX1 | 47 | 7.454 | 0.1144 | Yes | ||
| 5 | PFDN2 | 74 | 6.806 | 0.1344 | Yes | ||
| 6 | HNRPA1 | 80 | 6.737 | 0.1553 | Yes | ||
| 7 | SHMT1 | 272 | 4.709 | 0.1597 | Yes | ||
| 8 | EIF4B | 291 | 4.621 | 0.1733 | Yes | ||
| 9 | PABPC4 | 378 | 4.228 | 0.1819 | Yes | ||
| 10 | PABPC1 | 419 | 4.086 | 0.1926 | Yes | ||
| 11 | NR1D1 | 421 | 4.076 | 0.2053 | Yes | ||
| 12 | DDX3X | 560 | 3.707 | 0.2095 | Yes | ||
| 13 | CTSF | 579 | 3.645 | 0.2200 | Yes | ||
| 14 | GTF2A1 | 621 | 3.547 | 0.2289 | Yes | ||
| 15 | BOK | 702 | 3.410 | 0.2353 | Yes | ||
| 16 | MRPL27 | 751 | 3.320 | 0.2431 | Yes | ||
| 17 | EIF4E | 869 | 3.123 | 0.2466 | Yes | ||
| 18 | UTP14A | 953 | 2.985 | 0.2515 | Yes | ||
| 19 | AKAP1 | 1070 | 2.841 | 0.2541 | Yes | ||
| 20 | SYNCRIP | 1090 | 2.817 | 0.2619 | Yes | ||
| 21 | HEXA | 1207 | 2.652 | 0.2640 | Yes | ||
| 22 | SSR1 | 1245 | 2.606 | 0.2702 | Yes | ||
| 23 | FKBP11 | 1351 | 2.494 | 0.2723 | Yes | ||
| 24 | EIF3S10 | 1365 | 2.478 | 0.2794 | Yes | ||
| 25 | TIMM8A | 1368 | 2.475 | 0.2871 | Yes | ||
| 26 | RAB3IL1 | 1514 | 2.334 | 0.2865 | Yes | ||
| 27 | IRS4 | 1568 | 2.290 | 0.2909 | Yes | ||
| 28 | PER1 | 1581 | 2.278 | 0.2974 | Yes | ||
| 29 | HTATIP | 1672 | 2.218 | 0.2995 | Yes | ||
| 30 | KRTCAP2 | 1711 | 2.189 | 0.3043 | Yes | ||
| 31 | CCAR1 | 1883 | 2.075 | 0.3015 | Yes | ||
| 32 | PPRC1 | 1892 | 2.070 | 0.3076 | Yes | ||
| 33 | OPRS1 | 2136 | 1.935 | 0.3005 | Yes | ||
| 34 | HOXB4 | 2137 | 1.934 | 0.3066 | Yes | ||
| 35 | QTRTD1 | 2146 | 1.928 | 0.3122 | Yes | ||
| 36 | ATF4 | 2184 | 1.910 | 0.3162 | Yes | ||
| 37 | GADD45G | 2212 | 1.894 | 0.3207 | Yes | ||
| 38 | SNX16 | 2213 | 1.894 | 0.3267 | Yes | ||
| 39 | LIG3 | 2219 | 1.890 | 0.3323 | Yes | ||
| 40 | PTGES2 | 2285 | 1.852 | 0.3346 | Yes | ||
| 41 | SYT3 | 2348 | 1.827 | 0.3370 | Yes | ||
| 42 | RBBP4 | 2393 | 1.803 | 0.3403 | Yes | ||
| 43 | TOM1 | 2428 | 1.786 | 0.3441 | Yes | ||
| 44 | UBXD3 | 2601 | 1.714 | 0.3401 | Yes | ||
| 45 | STMN1 | 2649 | 1.690 | 0.3429 | Yes | ||
| 46 | TEF | 2673 | 1.680 | 0.3469 | Yes | ||
| 47 | CD164 | 3024 | 1.535 | 0.3328 | No | ||
| 48 | DRD1 | 3085 | 1.516 | 0.3343 | No | ||
| 49 | CHD4 | 3237 | 1.460 | 0.3307 | No | ||
| 50 | CELSR3 | 3283 | 1.444 | 0.3328 | No | ||
| 51 | EIF4G1 | 3294 | 1.441 | 0.3368 | No | ||
| 52 | COMMD3 | 3444 | 1.382 | 0.3330 | No | ||
| 53 | ALDH3B1 | 3522 | 1.358 | 0.3331 | No | ||
| 54 | SOCS5 | 3558 | 1.346 | 0.3355 | No | ||
| 55 | COPZ1 | 3731 | 1.288 | 0.3302 | No | ||
| 56 | SYT6 | 3793 | 1.270 | 0.3309 | No | ||
| 57 | UBE4B | 3801 | 1.267 | 0.3345 | No | ||
| 58 | PRPS1 | 4009 | 1.203 | 0.3270 | No | ||
| 59 | ATP6V1C1 | 4046 | 1.192 | 0.3288 | No | ||
| 60 | FBXL19 | 4199 | 1.153 | 0.3242 | No | ||
| 61 | GPC3 | 4315 | 1.125 | 0.3215 | No | ||
| 62 | CDC14A | 4380 | 1.107 | 0.3215 | No | ||
| 63 | ZCCHC7 | 4584 | 1.059 | 0.3138 | No | ||
| 64 | CDKN2C | 4817 | 1.002 | 0.3044 | No | ||
| 65 | ADAM12 | 4868 | 0.989 | 0.3048 | No | ||
| 66 | GGN | 5009 | 0.955 | 0.3002 | No | ||
| 67 | TOPORS | 5111 | 0.932 | 0.2977 | No | ||
| 68 | TAF6L | 5153 | 0.923 | 0.2983 | No | ||
| 69 | ZIC1 | 5244 | 0.898 | 0.2963 | No | ||
| 70 | AP1S2 | 5265 | 0.892 | 0.2980 | No | ||
| 71 | HOXA9 | 5376 | 0.866 | 0.2948 | No | ||
| 72 | MAT2A | 5468 | 0.847 | 0.2925 | No | ||
| 73 | RPL13A | 5528 | 0.833 | 0.2919 | No | ||
| 74 | GJA1 | 5530 | 0.833 | 0.2945 | No | ||
| 75 | ANKRD17 | 5746 | 0.790 | 0.2853 | No | ||
| 76 | NEUROD2 | 5814 | 0.776 | 0.2841 | No | ||
| 77 | XPO1 | 5864 | 0.766 | 0.2839 | No | ||
| 78 | MCM2 | 6089 | 0.721 | 0.2740 | No | ||
| 79 | PLCG2 | 6130 | 0.712 | 0.2741 | No | ||
| 80 | UTY | 6209 | 0.697 | 0.2720 | No | ||
| 81 | DNMT3A | 6216 | 0.696 | 0.2739 | No | ||
| 82 | MYST3 | 6354 | 0.664 | 0.2685 | No | ||
| 83 | H3F3A | 6649 | 0.605 | 0.2545 | No | ||
| 84 | PICALM | 6681 | 0.598 | 0.2547 | No | ||
| 85 | HOXD10 | 6913 | 0.548 | 0.2439 | No | ||
| 86 | PCDHA10 | 7146 | 0.502 | 0.2329 | No | ||
| 87 | HOXC5 | 7170 | 0.497 | 0.2332 | No | ||
| 88 | PTMA | 7233 | 0.486 | 0.2314 | No | ||
| 89 | TBC1D15 | 7493 | 0.434 | 0.2187 | No | ||
| 90 | HNRPD | 7882 | 0.358 | 0.1988 | No | ||
| 91 | UTX | 7962 | 0.343 | 0.1956 | No | ||
| 92 | LIN28 | 8035 | 0.329 | 0.1927 | No | ||
| 93 | SIRT1 | 8112 | 0.315 | 0.1896 | No | ||
| 94 | ALDH6A1 | 8915 | 0.170 | 0.1466 | No | ||
| 95 | NPM1 | 8937 | 0.166 | 0.1460 | No | ||
| 96 | NPTX1 | 8966 | 0.160 | 0.1450 | No | ||
| 97 | SLC1A7 | 8982 | 0.157 | 0.1447 | No | ||
| 98 | ARMET | 9257 | 0.106 | 0.1302 | No | ||
| 99 | SLC38A5 | 9503 | 0.055 | 0.1170 | No | ||
| 100 | ODC1 | 9549 | 0.045 | 0.1147 | No | ||
| 101 | TRIM37 | 9591 | 0.037 | 0.1126 | No | ||
| 102 | CPT1A | 9640 | 0.023 | 0.1101 | No | ||
| 103 | TRIB1 | 9842 | -0.020 | 0.0993 | No | ||
| 104 | DLX2 | 9916 | -0.037 | 0.0954 | No | ||
| 105 | CUL5 | 9919 | -0.039 | 0.0954 | No | ||
| 106 | HOXA11 | 9997 | -0.058 | 0.0915 | No | ||
| 107 | TNRC15 | 10018 | -0.063 | 0.0906 | No | ||
| 108 | FMR1 | 10231 | -0.107 | 0.0794 | No | ||
| 109 | UCHL1 | 10272 | -0.118 | 0.0776 | No | ||
| 110 | BMP6 | 10289 | -0.123 | 0.0771 | No | ||
| 111 | NEUROD1 | 10311 | -0.126 | 0.0764 | No | ||
| 112 | OPRD1 | 10533 | -0.175 | 0.0650 | No | ||
| 113 | TESK2 | 10584 | -0.183 | 0.0628 | No | ||
| 114 | LAMP1 | 10676 | -0.201 | 0.0585 | No | ||
| 115 | POU3F2 | 10794 | -0.225 | 0.0529 | No | ||
| 116 | MEOX2 | 11127 | -0.296 | 0.0358 | No | ||
| 117 | PAX6 | 11207 | -0.311 | 0.0325 | No | ||
| 118 | TGFB2 | 11211 | -0.313 | 0.0333 | No | ||
| 119 | CBARA1 | 11231 | -0.316 | 0.0333 | No | ||
| 120 | SNX5 | 11340 | -0.339 | 0.0285 | No | ||
| 121 | RGL1 | 11586 | -0.392 | 0.0165 | No | ||
| 122 | USP15 | 11955 | -0.475 | -0.0020 | No | ||
| 123 | NFX1 | 11979 | -0.480 | -0.0017 | No | ||
| 124 | B3GALT2 | 12101 | -0.506 | -0.0067 | No | ||
| 125 | RPS28 | 12184 | -0.526 | -0.0095 | No | ||
| 126 | TGIF2 | 12299 | -0.556 | -0.0139 | No | ||
| 127 | NDUFA7 | 12306 | -0.558 | -0.0125 | No | ||
| 128 | GATA5 | 12320 | -0.562 | -0.0114 | No | ||
| 129 | SNCAIP | 12452 | -0.593 | -0.0167 | No | ||
| 130 | HOXB5 | 12480 | -0.599 | -0.0163 | No | ||
| 131 | ELAVL3 | 12621 | -0.632 | -0.0219 | No | ||
| 132 | SEPT3 | 12680 | -0.647 | -0.0230 | No | ||
| 133 | RNF146 | 12775 | -0.671 | -0.0260 | No | ||
| 134 | SLCO1C1 | 12854 | -0.692 | -0.0280 | No | ||
| 135 | PPM1A | 12935 | -0.712 | -0.0301 | No | ||
| 136 | DCTN4 | 13091 | -0.757 | -0.0361 | No | ||
| 137 | CHST11 | 13110 | -0.763 | -0.0347 | No | ||
| 138 | NR0B2 | 13116 | -0.765 | -0.0326 | No | ||
| 139 | HOXA1 | 13771 | -0.955 | -0.0650 | No | ||
| 140 | RPA1 | 13806 | -0.965 | -0.0638 | No | ||
| 141 | SEC23IP | 14031 | -1.042 | -0.0727 | No | ||
| 142 | ILF3 | 14335 | -1.146 | -0.0855 | No | ||
| 143 | TAC1 | 14458 | -1.189 | -0.0884 | No | ||
| 144 | DUSP7 | 14571 | -1.226 | -0.0906 | No | ||
| 145 | NRIP3 | 14637 | -1.250 | -0.0902 | No | ||
| 146 | SCYL1 | 14786 | -1.309 | -0.0941 | No | ||
| 147 | ZNF318 | 14915 | -1.364 | -0.0968 | No | ||
| 148 | PLAGL1 | 14924 | -1.368 | -0.0929 | No | ||
| 149 | CITED2 | 15581 | -1.686 | -0.1232 | No | ||
| 150 | UBE2B | 15649 | -1.725 | -0.1214 | No | ||
| 151 | CBX5 | 15705 | -1.755 | -0.1189 | No | ||
| 152 | HOXB7 | 15886 | -1.873 | -0.1227 | No | ||
| 153 | SMYD4 | 16061 | -1.973 | -0.1260 | No | ||
| 154 | VPS33A | 16071 | -1.983 | -0.1202 | No | ||
| 155 | ARPC5 | 16190 | -2.087 | -0.1201 | No | ||
| 156 | LEF1 | 16397 | -2.266 | -0.1241 | No | ||
| 157 | NIT1 | 16577 | -2.439 | -0.1262 | No | ||
| 158 | RELB | 16614 | -2.472 | -0.1203 | No | ||
| 159 | UBXD1 | 16794 | -2.674 | -0.1216 | No | ||
| 160 | IGF2R | 16943 | -2.855 | -0.1207 | No | ||
| 161 | SLC38A2 | 16997 | -2.927 | -0.1144 | No | ||
| 162 | KCNK5 | 17088 | -3.050 | -0.1097 | No | ||
| 163 | VPS16 | 17345 | -3.485 | -0.1126 | No | ||
| 164 | GNAS | 17496 | -3.817 | -0.1087 | No | ||
| 165 | CBX8 | 17589 | -3.985 | -0.1012 | No | ||
| 166 | GABARAP | 17603 | -4.008 | -0.0893 | No | ||
| 167 | CUGBP1 | 17752 | -4.386 | -0.0835 | No | ||
| 168 | EPC1 | 17925 | -4.900 | -0.0775 | No | ||
| 169 | KBTBD2 | 18041 | -5.284 | -0.0671 | No | ||
| 170 | BRD2 | 18095 | -5.480 | -0.0527 | No | ||
| 171 | ISGF3G | 18468 | -8.565 | -0.0460 | No | ||
| 172 | HOXA7 | 18589 | -17.167 | 0.0015 | No |
