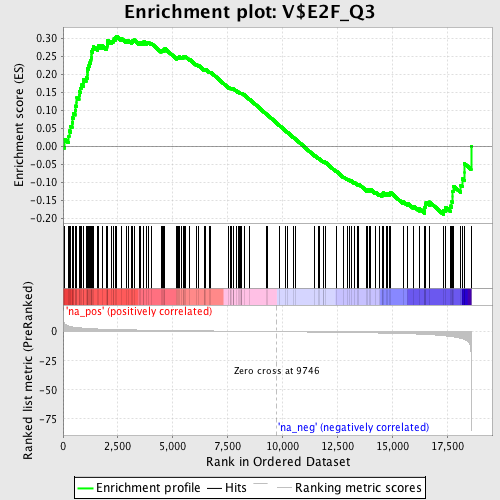
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_Q3 |
| Enrichment Score (ES) | 0.30583695 |
| Normalized Enrichment Score (NES) | 1.3322221 |
| Nominal p-value | 0.006369427 |
| FDR q-value | 0.21428445 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HNRPA1 | 80 | 6.737 | 0.0197 | Yes | ||
| 2 | IMPDH2 | 260 | 4.782 | 0.0270 | Yes | ||
| 3 | SHMT1 | 272 | 4.709 | 0.0432 | Yes | ||
| 4 | SLC38A1 | 347 | 4.333 | 0.0547 | Yes | ||
| 5 | TCP1 | 410 | 4.102 | 0.0659 | Yes | ||
| 6 | PVRL1 | 429 | 4.049 | 0.0794 | Yes | ||
| 7 | PCSK4 | 463 | 3.934 | 0.0916 | Yes | ||
| 8 | ATAD2 | 544 | 3.756 | 0.1007 | Yes | ||
| 9 | UNG | 571 | 3.670 | 0.1123 | Yes | ||
| 10 | USP1 | 625 | 3.544 | 0.1221 | Yes | ||
| 11 | ABCF2 | 627 | 3.544 | 0.1347 | Yes | ||
| 12 | EPHB2 | 726 | 3.367 | 0.1414 | Yes | ||
| 13 | SLC25A3 | 752 | 3.317 | 0.1518 | Yes | ||
| 14 | AP4M1 | 796 | 3.247 | 0.1611 | Yes | ||
| 15 | MCM6 | 829 | 3.191 | 0.1707 | Yes | ||
| 16 | DNAJC11 | 916 | 3.048 | 0.1769 | Yes | ||
| 17 | PPP1R8 | 938 | 3.014 | 0.1865 | Yes | ||
| 18 | RFC1 | 1047 | 2.871 | 0.1909 | Yes | ||
| 19 | RANBP1 | 1098 | 2.805 | 0.1982 | Yes | ||
| 20 | LIG1 | 1101 | 2.799 | 0.2081 | Yes | ||
| 21 | TFRC | 1111 | 2.779 | 0.2175 | Yes | ||
| 22 | NCL | 1174 | 2.702 | 0.2238 | Yes | ||
| 23 | RAD51 | 1206 | 2.652 | 0.2316 | Yes | ||
| 24 | E2F1 | 1260 | 2.582 | 0.2379 | Yes | ||
| 25 | NASP | 1272 | 2.565 | 0.2464 | Yes | ||
| 26 | RBBP7 | 1300 | 2.547 | 0.2541 | Yes | ||
| 27 | UXT | 1302 | 2.546 | 0.2631 | Yes | ||
| 28 | MCM4 | 1339 | 2.506 | 0.2701 | Yes | ||
| 29 | ING3 | 1377 | 2.470 | 0.2769 | Yes | ||
| 30 | KLF4 | 1583 | 2.277 | 0.2739 | Yes | ||
| 31 | GCH1 | 1625 | 2.251 | 0.2797 | Yes | ||
| 32 | E2F3 | 1780 | 2.138 | 0.2790 | Yes | ||
| 33 | APRIN | 1975 | 2.026 | 0.2757 | Yes | ||
| 34 | MCM3 | 2001 | 2.009 | 0.2815 | Yes | ||
| 35 | ADAMTS2 | 2026 | 1.998 | 0.2873 | Yes | ||
| 36 | MCM7 | 2035 | 1.992 | 0.2940 | Yes | ||
| 37 | POLA2 | 2218 | 1.890 | 0.2909 | Yes | ||
| 38 | OLFML3 | 2277 | 1.854 | 0.2943 | Yes | ||
| 39 | PIAS1 | 2318 | 1.841 | 0.2987 | Yes | ||
| 40 | RBBP4 | 2393 | 1.803 | 0.3011 | Yes | ||
| 41 | NRP2 | 2425 | 1.787 | 0.3058 | Yes | ||
| 42 | STMN1 | 2649 | 1.690 | 0.2998 | No | ||
| 43 | RHD | 2878 | 1.593 | 0.2931 | No | ||
| 44 | PCSK1 | 2959 | 1.560 | 0.2944 | No | ||
| 45 | KCNA1 | 3120 | 1.503 | 0.2910 | No | ||
| 46 | SFRS2 | 3178 | 1.480 | 0.2932 | No | ||
| 47 | CDC25A | 3235 | 1.463 | 0.2954 | No | ||
| 48 | PHF15 | 3471 | 1.374 | 0.2876 | No | ||
| 49 | TBX3 | 3544 | 1.353 | 0.2885 | No | ||
| 50 | PLK4 | 3662 | 1.309 | 0.2868 | No | ||
| 51 | HTF9C | 3683 | 1.304 | 0.2904 | No | ||
| 52 | KPNB1 | 3811 | 1.264 | 0.2880 | No | ||
| 53 | HMGN2 | 3884 | 1.241 | 0.2886 | No | ||
| 54 | PRPS1 | 4009 | 1.203 | 0.2861 | No | ||
| 55 | GMNN | 4463 | 1.085 | 0.2655 | No | ||
| 56 | ARHGAP6 | 4528 | 1.070 | 0.2658 | No | ||
| 57 | DLST | 4566 | 1.062 | 0.2676 | No | ||
| 58 | CDC6 | 4627 | 1.049 | 0.2681 | No | ||
| 59 | LASP1 | 4638 | 1.048 | 0.2713 | No | ||
| 60 | CDCA7 | 5180 | 0.919 | 0.2453 | No | ||
| 61 | HS6ST3 | 5217 | 0.906 | 0.2466 | No | ||
| 62 | AP1S2 | 5265 | 0.892 | 0.2472 | No | ||
| 63 | ADCY8 | 5288 | 0.886 | 0.2492 | No | ||
| 64 | FMO4 | 5406 | 0.860 | 0.2459 | No | ||
| 65 | MAT2A | 5468 | 0.847 | 0.2456 | No | ||
| 66 | BAT3 | 5473 | 0.846 | 0.2484 | No | ||
| 67 | TRIM47 | 5527 | 0.833 | 0.2485 | No | ||
| 68 | SLC6A4 | 5579 | 0.826 | 0.2487 | No | ||
| 69 | ASCL1 | 5781 | 0.783 | 0.2406 | No | ||
| 70 | MCM2 | 6089 | 0.721 | 0.2265 | No | ||
| 71 | PAQR4 | 6181 | 0.701 | 0.2241 | No | ||
| 72 | HIST1H2BK | 6424 | 0.650 | 0.2133 | No | ||
| 73 | STAG2 | 6458 | 0.642 | 0.2138 | No | ||
| 74 | BARHL1 | 6511 | 0.631 | 0.2133 | No | ||
| 75 | KCND2 | 6680 | 0.598 | 0.2063 | No | ||
| 76 | POLE2 | 6724 | 0.587 | 0.2061 | No | ||
| 77 | PODN | 7546 | 0.426 | 0.1631 | No | ||
| 78 | CPNE5 | 7618 | 0.410 | 0.1608 | No | ||
| 79 | JPH1 | 7623 | 0.409 | 0.1620 | No | ||
| 80 | SNPH | 7667 | 0.402 | 0.1611 | No | ||
| 81 | SEMA5A | 7751 | 0.386 | 0.1580 | No | ||
| 82 | MRPL18 | 7759 | 0.385 | 0.1590 | No | ||
| 83 | GRIK2 | 7771 | 0.383 | 0.1597 | No | ||
| 84 | HNRPD | 7882 | 0.358 | 0.1551 | No | ||
| 85 | STK35 | 7977 | 0.339 | 0.1512 | No | ||
| 86 | SFRS1 | 8028 | 0.331 | 0.1497 | No | ||
| 87 | SMAD6 | 8080 | 0.321 | 0.1480 | No | ||
| 88 | UFD1L | 8109 | 0.315 | 0.1476 | No | ||
| 89 | H2AFZ | 8134 | 0.311 | 0.1475 | No | ||
| 90 | CTCF | 8141 | 0.310 | 0.1482 | No | ||
| 91 | NDUFA11 | 8249 | 0.292 | 0.1435 | No | ||
| 92 | WEE1 | 8488 | 0.246 | 0.1315 | No | ||
| 93 | PAPOLG | 9268 | 0.104 | 0.0897 | No | ||
| 94 | NPR3 | 9316 | 0.093 | 0.0875 | No | ||
| 95 | TOPBP1 | 9864 | -0.025 | 0.0579 | No | ||
| 96 | ERBB2IP | 10144 | -0.089 | 0.0431 | No | ||
| 97 | HIST1H1D | 10232 | -0.107 | 0.0388 | No | ||
| 98 | SEZ6 | 10478 | -0.164 | 0.0261 | No | ||
| 99 | CLTA | 10603 | -0.187 | 0.0201 | No | ||
| 100 | DNMT1 | 11467 | -0.366 | -0.0253 | No | ||
| 101 | DMD | 11628 | -0.401 | -0.0326 | No | ||
| 102 | IPO7 | 11662 | -0.410 | -0.0329 | No | ||
| 103 | FBXO5 | 11851 | -0.451 | -0.0415 | No | ||
| 104 | EPHB1 | 11889 | -0.462 | -0.0418 | No | ||
| 105 | ONECUT1 | 11966 | -0.478 | -0.0442 | No | ||
| 106 | CASP8AP2 | 12448 | -0.592 | -0.0682 | No | ||
| 107 | NUP153 | 12795 | -0.676 | -0.0845 | No | ||
| 108 | NOL4 | 12946 | -0.714 | -0.0901 | No | ||
| 109 | ARID4A | 13045 | -0.742 | -0.0927 | No | ||
| 110 | PCNA | 13131 | -0.768 | -0.0946 | No | ||
| 111 | SALL1 | 13282 | -0.805 | -0.0998 | No | ||
| 112 | HIST1H2AH | 13415 | -0.843 | -0.1040 | No | ||
| 113 | ATE1 | 13483 | -0.864 | -0.1045 | No | ||
| 114 | EZH2 | 13846 | -0.978 | -0.1206 | No | ||
| 115 | TREX2 | 13869 | -0.984 | -0.1183 | No | ||
| 116 | PEG3 | 13972 | -1.019 | -0.1202 | No | ||
| 117 | CACNA1G | 13997 | -1.029 | -0.1179 | No | ||
| 118 | SSBP3 | 14229 | -1.112 | -0.1264 | No | ||
| 119 | MEIS1 | 14412 | -1.172 | -0.1321 | No | ||
| 120 | MDGA1 | 14536 | -1.215 | -0.1344 | No | ||
| 121 | PRKDC | 14560 | -1.225 | -0.1313 | No | ||
| 122 | ORC1L | 14579 | -1.228 | -0.1279 | No | ||
| 123 | FLI1 | 14749 | -1.297 | -0.1324 | No | ||
| 124 | CDC45L | 14785 | -1.309 | -0.1296 | No | ||
| 125 | ATRX | 14883 | -1.351 | -0.1301 | No | ||
| 126 | LHX5 | 14908 | -1.362 | -0.1265 | No | ||
| 127 | MCM8 | 15507 | -1.647 | -0.1530 | No | ||
| 128 | CBX5 | 15705 | -1.755 | -0.1574 | No | ||
| 129 | ZCCHC8 | 15971 | -1.916 | -0.1649 | No | ||
| 130 | PPP1CC | 16230 | -2.120 | -0.1714 | No | ||
| 131 | CTDSP1 | 16478 | -2.345 | -0.1764 | No | ||
| 132 | MAZ | 16484 | -2.346 | -0.1683 | No | ||
| 133 | MXD3 | 16506 | -2.368 | -0.1610 | No | ||
| 134 | CNOT3 | 16537 | -2.399 | -0.1541 | No | ||
| 135 | SLC25A14 | 16698 | -2.559 | -0.1536 | No | ||
| 136 | PHF13 | 17340 | -3.474 | -0.1759 | No | ||
| 137 | KLF5 | 17429 | -3.641 | -0.1677 | No | ||
| 138 | INSM1 | 17646 | -4.125 | -0.1647 | No | ||
| 139 | SYT11 | 17717 | -4.305 | -0.1532 | No | ||
| 140 | PHF12 | 17760 | -4.415 | -0.1397 | No | ||
| 141 | PIM1 | 17761 | -4.415 | -0.1240 | No | ||
| 142 | DPYSL2 | 17791 | -4.509 | -0.1095 | No | ||
| 143 | E2F7 | 18127 | -5.625 | -0.1075 | No | ||
| 144 | RAB11B | 18190 | -5.975 | -0.0896 | No | ||
| 145 | DCK | 18298 | -6.679 | -0.0716 | No | ||
| 146 | DDB2 | 18301 | -6.705 | -0.0478 | No | ||
| 147 | TBX6 | 18594 | -18.181 | 0.0012 | No |
